Single-cell sequencing has emerged as a novel method for providing a deeper insight into the genetic information pertaining to the genome, transcriptome, exome, and other single-cell multi-omics. This type of sequencing has revolutionized the field of biomedical research and medicine with its ability to provide a precise understanding of the inner workings of cells with in-depth coverage of genetic sequences and their association with disease status.
Image Credit: Yurchanka Siarhei/Shutterstock.com
The comprehensive analysis of the genome or parts of the genome enables the detection of somatic mutations, such as single nucleotide variants, copy number variants, insertions and deletions, and gene fusions. Cell-to-cell heterogeneity is ubiquitous among cancer cells. Analyzing this may not be possible through conventional bulk-cell sequencing methods, so utilizing single-cell sequencing can have prominent advantages for patient treatment.
Single Cell Sequencing
Single-cell sequencing comprises deep sequencing of individual cells through sequencing methods such as next-generation sequencing (NGS); this enables further insight into the genetic information and the function of cells in their microenvironment.
This type of sequencing is considered superior to conventional sequencing methods that are limited to the average of many cells, have a small analysis level, and are ineffective in collecting information on cellular heterogeneity.
The principle of this method comprises isolating a single cell for sequencing, and through DNA or RNA extraction, the cell can be examined more precisely for the genomic, transcriptomic, or epigenetic information. This can be visualized in Figure 1, which outlines the process.
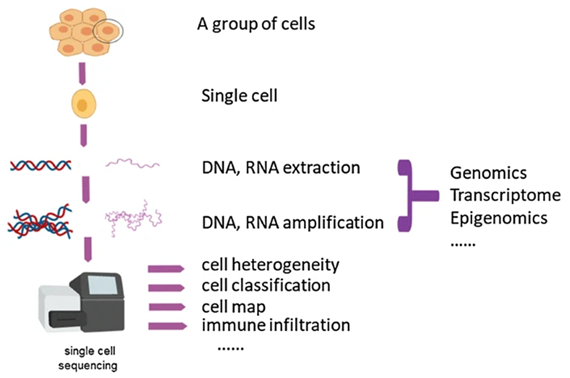
Figure 1 demonstrates the process of single-cell sequencing and the how single cells can be examined with deep coverage of the genome, transcriptome, and even for epigenetic purposes. Available at: The single-cell sequencing: new developments and medical applications | Cell & Bioscience | Full Text (biomedcentral.com)
As this process has advanced, the cost of single-cell sequencing has decreased and has been applied to various fields and applications, such as tumors, microbiology, neurology, reproduction, and immunity.
Benefits of Single-Cell Sequencing
Through this approach, the deep analysis of information from cells can ensure genomic alterations are not undetected. The lack of detection of these critical mutations can result in ineffective drug treatment, which may lead to disease progression.
The benefit of using single-cell sequencing, which allows for in-depth coverage of sequences and nucleotides, enables these mutations to be identified, and this can help guide patient treatment to ensure the correct drug is provided, catering to any resistance mechanisms characterizing the patient’s cancer.
This type of sequencing can advance the field of precision or personalized medicine, as it focuses deeply on the patient and their genomic alterations, rather than a broad universal ‘one-size-fits-all’ use of conventional chemotherapy drugs. This type of sequencing can further ensure a patient is provided with optimum care directly targeted to their cancer.
Targeting Mutations
An example of this includes lung cancer, which would require three classes of targets, (i) ‘must-test’ biomarkers, i.e., epidermal growth factor receptor, (ii) ‘should-test’ biomarkers, i.e., receptor tyrosine kinase MET proto-oncogene and (iii) investigative biomarkers.
Patients with advanced lung adenocarcinoma (ADC), a type of non-small lung cancer, can have various mutations such as anaplastic lymphoma kinase (ALK), reactive oxygen species (ROS) proto-oncogene 1 (ROS1), rearranged during transfection proto-oncogene (RET), and neurotrophin kinase (NTRK).
Detecting these gene rearrangements is critical as these require treatment with tyrosine kinase inhibitors, and if treated with a drug that does not target these mutations specifically, the disease status of cancer may be prolonged and can become severe.
Utilizing sequencing for diagnostic purposes to guide clinical decisions in this manner can be critical for a patient who can either be provided with targeted treatment with a good prognosis of their cancer or be faced with secondary malignancies with metastasis, which could ultimately result in lower prognoses and a possible increase in mortality.
Lung cancer is the leading global cause of mortality and is responsible for 1.4 million deaths yearly. While there have been advancements in cancer research, the overall five-year survival has only slightly increased over 2.5 decades, with a rate of 16%. With most patients being diagnosed at a late stage and a lack of early diagnostic procedures for certain lung cancers, such as non-small lung cancer, curative treatment at this stage is challenging.
This illustrates the requirement for advancements in this field as 80-85% of diagnoses are for non-small lung cancer, and the lack of early diagnosis and treatments at a late stage is very concerning.
Translational Significance
Single-cell sequencing, both DNA and RNA, may be a component of advancing precision medicine for the future of cancer diagnostics to provide a comprehensive analysis of cells and ensure the highest characterization of patient disorders. Early and accurate diagnosis with precise insight into genomic alterations can further comprehend the most effective treatment plan.
This strategy may enable better prognoses and reduce mortality rates globally. The field of precision medicine in this manner may have some challenges for use in a clinical setting. However, with the use of single-cell sequencing for not only cancer but other disorders such as Alzheimer’s and other infectious diseases, the burden on the healthcare system may be alleviated.
Sources:
- Huang, C., Ju, D., Chang, C., Muralidhar Reddy, P. and Velmurugan, B., 2017. A review on the effects of current chemotherapy drugs and natural agents in treating non–small cell lung cancer. BioMedicine, 7(4), p.23. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5682982/
- Bruno, R. and Fontanini, G., 2020. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics, 10(8), p.521. Available at: https://pubmed.ncbi.nlm.nih.gov/32726941/
- Vu, T., Nguyen, H., Calza, S., Kalari, K., Wang, L. and Pawitan, Y., 2019. Cell-level somatic mutation detection from single-cell RNA sequencing. Bioinformatics, 35(22), pp.4679-4687. Available at: https://pubmed.ncbi.nlm.nih.gov/31028395/
- Gawad, C., Koh, W. & Quake, S. Single-cell genome sequencing: current state of the science. Nat Rev Genet 17, 175–188 (2016). https://doi.org/10.1038/nrg.2015.16
- Tang, X., Huang, Y., Lei, J. et al. The single-cell sequencing: new developments and medical applications. Cell Biosci 9, 53 (2019). https://doi.org/10.1186/s13578-019-0314-y
- Evrony, G., Hinch, A. and Luo, C., 2021. Applications of Single-Cell DNA Sequencing. Annual Review of Genomics and Human Genetics, 22(1), pp.171-197. Available at: https://pubmed.ncbi.nlm.nih.gov/33722077/
Further Reading