DNA is made up of nucleobases, which are depicted by the letters G, A, T, and C. They are the foundation of the genetic code and are found in all living organisms. However, another base, indicated by the letter Z occurs in a bacteriophage.
A : T, G : C and Z : T bonds. Image Credit: © Adobe Stock.
This one-of-a-kind occurrence, the only one witnessed to date, has long been a mystery. The biosynthesis pathway of this base has now been illustrated by scientists from the Institut Pasteur and the CNRS, in cooperation with the CEA. This study was published in the 2021 issue of Science journal.
DNA, also known as deoxyribonucleic acid, is a molecule that acts as the medium for all living beings to store genetic information. That is a double helix differentiated by alternating purine nucleobases (guanine and adenine) and pyrimidine nucleobases (deoxycytidine and cytidine).
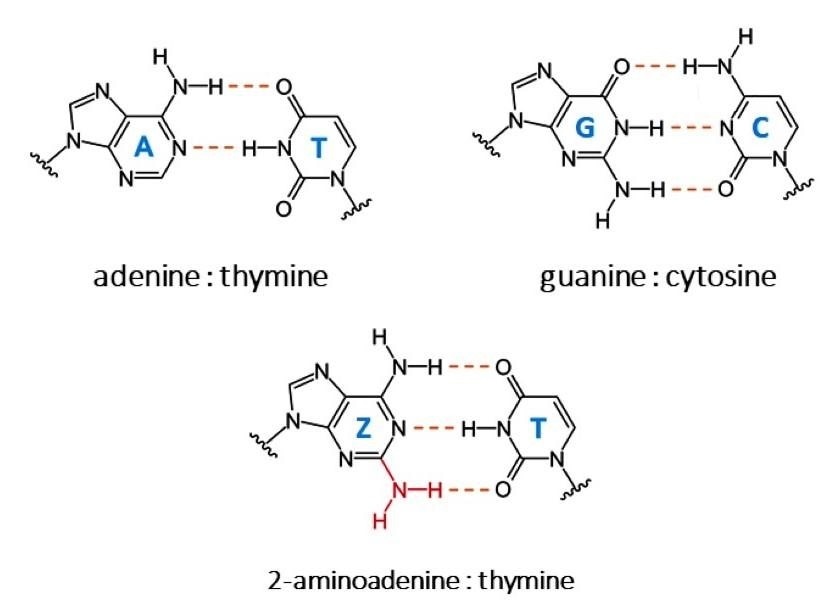
The bases of each DNA strand are bound together at the center of the helix, connecting the two DNA strands: adenine forms two hydrogen bonds with thymine (A–T), and guanine forms three hydrogen bonds with cytosine (G–C). For one exception, this extends to all living things.
Cyanophage S-2L, an exception to conventional genetics
Cyanophage S-2L is a bacteriophage (a virus that infects bacteria). Adenine is totally substituted by another base, 2-aminoadenine, in this phage (indicated by the letter Z). Instead of the regular two bonds between adenine and thymine, the latter forms three hydrogen bonds with thymine (Z–T).
This increased number of bonds improves the DNA’s stability at high temperatures while also changing its conformation, making the DNA less well known by small molecules and proteins.
2-aminoadenine biosynthesis pathway elucidated
Cyanophage S-2L has been the only known exception since its discovery in 1977, and the biosynthesis mechanism of 2-aminoadenine has remained elusive. In partnership with the CEA, scientists from the Institut Pasteur and the CNRS recently explained this biosynthesis mechanism and showed its enzymatic origins.
They accomplished this by discovering a homolog of the well-known enzyme succinoadenylate synthase (PurA) in the genome of the cyanophage S-2L. A phylogenetic study of this enzyme family showed a connection between the PurZ homolog and the PurA enzyme in archaea.
This suggests that the homolog is an ancient enzyme that most likely had an evolutionary benefit. The Crystallography Platform at the Institut Pasteur was used for the study.
The identification of the biosynthesis process and the latest Z–T base pair demonstrates that new bases can be inserted into genetic material enzymatically. This raises the number of coding bases in DNA, opening the way for synthetic genetic biopolymers to be created.
Source:
Journal reference:
Sleiman, D., et al. (2021) A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science. doi.org/10.1126/science.abe6494.