Researchers recently identified the means to get the best DNA to communicate with membranes in the body, leading the way for the development of “mini biological computers” in droplets that have significant uses in biosensing and mRNA vaccines.
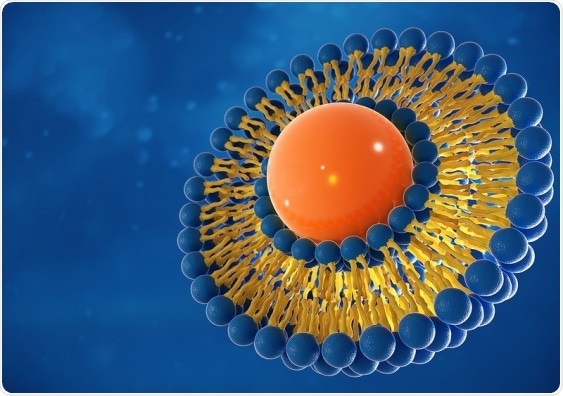
The study discovered the best way to design and build DNA “nanostructures” to effectively manipulate synthetic liposomes (pictured)—tiny bubbles which have traditionally been used to deliver drugs for cancer and other diseases. Image Credit: Shutterstock.
University of New South Wales’s (UNSW) Dr Matthew Baker and the University of Sydney’s Dr Shelley Wickham co-headed the research, which was published in the Nucleic Acids Research journal.
The study identified a better means to design and develop DNA “nanostructures” to efficiently control synthetic liposomes—tiny bubbles which were conventionally used to supply drugs for cancer and other diseases.
Greater applications like creating small molecular systems that sense their environment and respond to a signal to discharge cargo, like a drug molecule nearing its target, can be achieved by modifying the porosity, shape, and reactivity of liposomes.
Lead author Dr. Matt Baker from UNSW’s School of Biotechnology and Biomolecular Sciences remarks that the research identified the means to build “little blocks” out of DNA and figured out the better means to label the blocks with cholesterol to stick them to the lipids—the major constituents of animal and plant cells.
One major application of our study is biosensing: you could stick some droplets in a person or patient, as it moves through the body it records local environment, processes this and delivers a result so you can ‘read out’ the local environment.”
Dr Matt Baker, Study Lead Author, School of Biotechnology and Biomolecular Sciences, University of New South Wales
Liposome nanotechnology is now renowned due to the use of liposomes along with RNA vaccines like the Moderna and Pfizer COVID-19 vaccines.
Dr Baker adds, “This work shows new ways to corral liposomes into place and then pop them open at just the right time. What is better is because they are built from the bottom-up out of individual parts we design, we can easily bolt in and out different components to change the way they work.”
Researchers earlier struggled to identify the correct buffer conditions for lipids and liposomes to ensure that their DNA “computers” in fact adhered to liposomes.
The scientists also had difficulty figuring out the better means to decorate the DNA with cholesterols so that it can find the way to the membrane along with remaining there as long as needed.
Is it better at the edge? The center? Heaps of them? Few of them? Close as possible to structure, or far as possible? We looked at all these things and showed that we could make good conditions for DNA structures to bind to liposomes reliably and ‘do something’.”
Dr Matt Baker, Study Lead Author, School of Biotechnology and Biomolecular Sciences, University of New South Wales
Dr. Baker remarks that the membranes are crucial in life as they enable the formation of compartments and thereby the separation of various kinds of tissue and cells.
This all relies on membranes being generally quite impermeable. Here we have built new DNA nanotechnology where we can punch holes in membranes, on-demand, to be able to pass important signals across a membrane. This is ultimately the basis in life of how cells communicate with each other, and how something useful can be made in one cell and then exported to be used elsewhere.”
Dr Matt Baker, Study Lead Author, School of Biotechnology and Biomolecular Sciences, University of New South Wales
However, in pathogens, membranes can be interrupted to kill cells, or viruses can replicate by creeping into cells. The researchers further intend to carry on their research on how to control DNA-based pores that can be induced with light to develop synthetic retinas from completely new parts.
Source:
Journal reference:
Singh, J. K. D., et al. (2021) Binding of DNA origami to lipids: maximizing yield and switching via strand displacement. Nucleic Acids Research. doi.org/10.1093/nar/gkab888.