Since the passage of the DNA replication fork destroys nucleosomes on the DNA, chromatin inheritance during cell division requires the replication of DNA and the assembling of nucleosomes onto the duplicated DNA. Half of the histones for replication-coupled nucleosome assembly are derived from disrupted parental nucleosomes, while the other half is produced from scratch.
 Structures of CAF-1 and CAF-1 bound to histones H3-H4. Image Credit: Prof. XU Ruiming’s group
Structures of CAF-1 and CAF-1 bound to histones H3-H4. Image Credit: Prof. XU Ruiming’s group
CAF-1 is an evolutionarily conserved heterotrimeric protein complex that is in charge of the deposition of newly generated histones H3 and H4 onto DNA. The paucity of structural knowledge regarding CAF-1, on the other hand, has hampered comprehension of the molecular process underpinning de novo nucleosome assembly.
A research team led by Prof. Ruiming Xu from the Chinese Academy of Sciences’ Institute of Biophysics (IBP), in collaboration with Prof. Guohong Li, Prof. Bing Xhu, and Prof. Chaopei Liu, all from IBP, revealed high-resolution structures of CAF-1 and CAF-1 bound to histones H3-H4 in a study published in Science on August 25th, 2023.
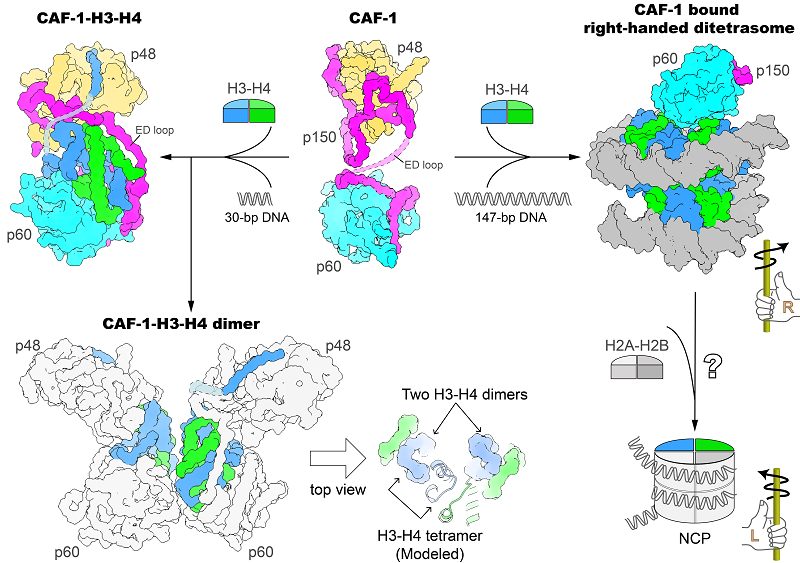
The researchers initially discovered the crystal structure of human CAF-1’s core domain in the absence of histones, followed by the crystal structure of CAF-1 in association with histones H3 and H4.

Image Credit: vitstudio/Shutterstock.com
According to the findings, a CAF-1 complex binds to an H3-H4 heterodimer mostly via the p60 subunit and the ED loop of the p150 subunit. The C-terminal region of the ED loop is critical to CAF-1's biological activity, as determined by in vitro histone binding and plasmid supercoiling tests and also by the in vivo nascent nucleosome assembly mapping and transcriptome investigations.
The CAF-1-H3-H4 combination dimerizes when a 30-bp DNA oligomer is present, and the researchers also determined the cryo-EM structure of a 2:2 CAF-1-H3-H4 complex.
This structure demonstrated that two adjacent H3-H4 heterodimers are geared for the formation of an H3-H4 tetramer through the dimerization of two H3s, but the positioning of the two heterodimers is not yet in the precise geometric arrangement of an H3-H4 tetramer.
This indicates that remodeling of histone H3-H4 binding by CAF-1, presumably with the binding of a longer DNA fragment, is required to assemble the H3-H4 tetramer, which is a crucial component of the nucleosome.
A right-handed di-tetrasome structure coupled to CAF-1 was seen by the researchers utilizing a 147-bp DNA fragment. This structure was further validated by analysis using the single-molecule freely orbiting magnetic tweezer (FOMT) approach at a physiological salt concentration.
This finding opened up a new avenue for mechanistic research into the nucleosome assembly process and suggested that a right-handed nucleosome precursor could be involved in replication-coupled nucleosome assembly.
Source:
Journal reference:
Liu, C.-P., et al. (2023). Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1. Science. doi.org/10.1126/science.add8673