Reviewed by Danielle Ellis, B.Sc.Nov 9 2023
Researchers from Kiel University (CAU) and the Max Planck Institute for Marine Microbiology in Bremen are shedding light on the enigmatic world of bacteria that coexist or parasitize animal hosts.
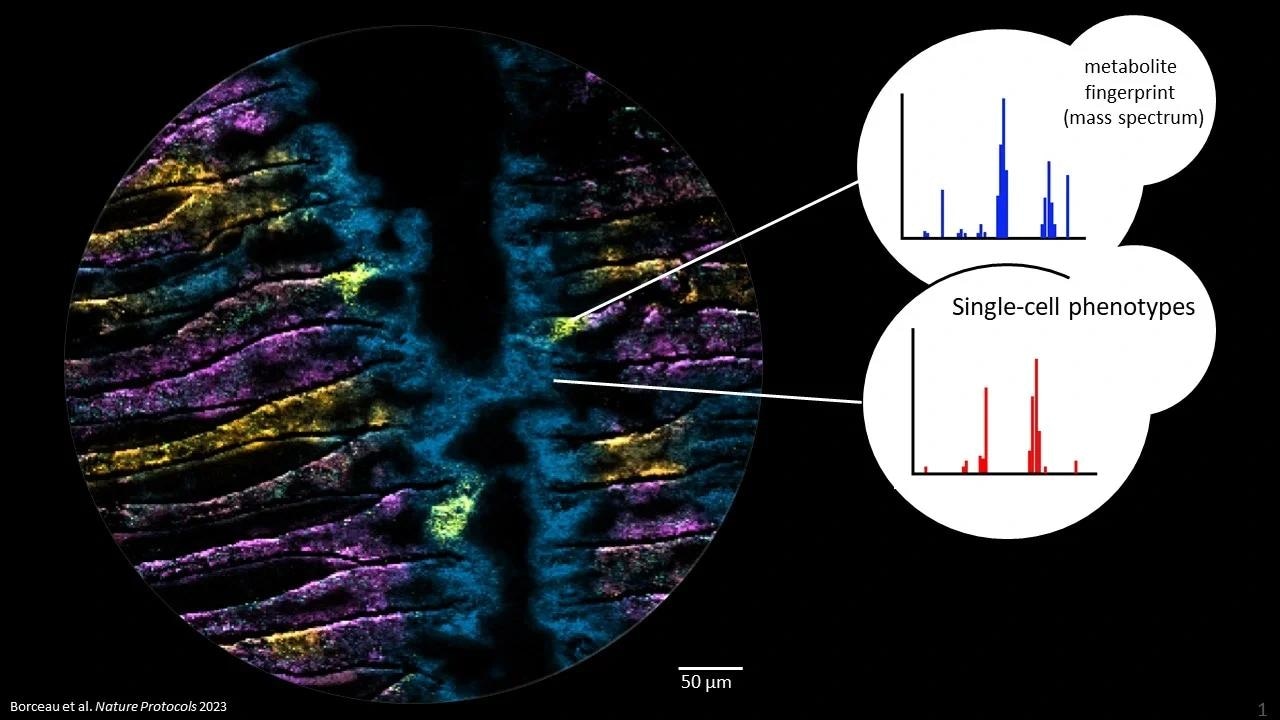 Metabolites in microbiomes on the micron scale. Each color in the picture represents a different metabolite, imaged with Mass spectrometry imaging at highest pixel resolution (2 um). The three yellow hotspots show epithelial cells infected with a bacterium and its metabolic consequences. Image Credit: Kiel University
Metabolites in microbiomes on the micron scale. Each color in the picture represents a different metabolite, imaged with Mass spectrometry imaging at highest pixel resolution (2 um). The three yellow hotspots show epithelial cells infected with a bacterium and its metabolic consequences. Image Credit: Kiel University
Led by Prof. Dr Manuel Liebeke, the team has made a groundbreaking discovery, unraveling the intricacies of microbial interactions with their hosts. Traditionally, studying these bacteria has been challenging due to the inability to culture them in the lab. Instead, researchers rely on genomic information from environmental samples, offering theoretical insights into microorganism metabolism.
However, a critical gap existed in understanding their real-world activities. To bridge this knowledge gap, scientists turned their attention to the metabolome of bacteria—the entirety of their metabolic processes, encompassing substances like sugars and fats.
In a pioneering effort, Liebeke’s team devised a method to identify individual bacteria and simultaneously discern the metabolites present in cells, all without the need for laboratory cultivation.
This innovative approach enables the examination of how bacteria thrive as symbiotic occupants, such as in mussels. The team scrutinized hundreds of metabolites within an area smaller than one square millimeter.
Their findings, published in the journal Nature Protocols in September, mark a significant leap forward in comprehending the intricate relationships between bacteria and their hosts.
Image Credit: Lightspring/Shutterstock.com
A Frozen Moment Enables Detailed Observation
We create a snapshot, so to speak, of the bacteria at work, exactly as they are active in their natural environment, particularly within an animal cell. And we can do that at an impressive resolution of a few micrometers, about ten times thinner than a human hair.”
Dr Manuel Liebeke, Head, Metabolomics Department, Faculty of Agricultural and Nutritional Sciences, Kiel University
This method boasts a unique aspect involving the utilization of flash-frozen tissue, sliced into razor-thin sections. Subsequently, researchers employ a specialized mass spectrometry technique known as MALDI-MS imaging to capture a detailed snapshot of the chemical compounds within the cells.
Yet, extracting accurate conclusions from the metabolite images hinges on the researchers' knowledge of the bacteria responsible for producing or utilizing them. To surpass this challenge, the team incorporates fluorescence in situ hybridization (FISH) to pinpoint and map the precise locations of individual bacterial cells within the sample.
Applying this method to host-microbe communities will give us many exciting new insights into chemical communication between organisms.”
Patric Bourceau, Study Lead Author, Max Planck Institute for Marine Microbiology
Bourceau spearheads the development of the protocol designed to implement this method. This groundbreaking endeavor not only ushers in new possibilities for delving into bacterial studies but also holds promise for future applications.
Originating from the Max Planck Institute in Bremen, Liebeke’s fresh research unit at the CAU is actively employing this method to explore the human gut microbiome and its impact on metabolism, potentially enhancing our comprehension of inflammatory bowel diseases.
The publication of a comprehensive protocol now opens the doors for researchers worldwide to apply this technique.
In essence, the integration of microscopy and metabolomics—dedicated to the study of metabolites—proffers insights into the functional and chemical dynamics of host-microbe interactions. The ongoing advancements in MALDI-MSI technology enable the visualization of microbial colonies, biofilms, individual eukaryotic cells, and even bacterial microcolonies.
As of today, MALDI-MSI technology stands on the cusp of providing images of individual bacterial cells. The protocol outlined here lays the foundation for analyzing and comprehending metabolic interactions at the microscopic level, down to the micrometer scale.
Source:
Journal reference:
Bourceau, P., et al. (2023) Visualization of metabolites and microbes at high spatial resolution using MALDI mass spectrometry imaging and in situ fluorescence labelling. Nature Protocols. doi.org/10.1038/s41596-023-00864-1.