Bacteria collaborate and share nutrients among generations when they form communities. Using a recently invented technique, researchers at the University of Basel have been able to demonstrate this for the first time. This novel method makes it possible to monitor gene expression both spatially and temporally as bacterial populations evolve.
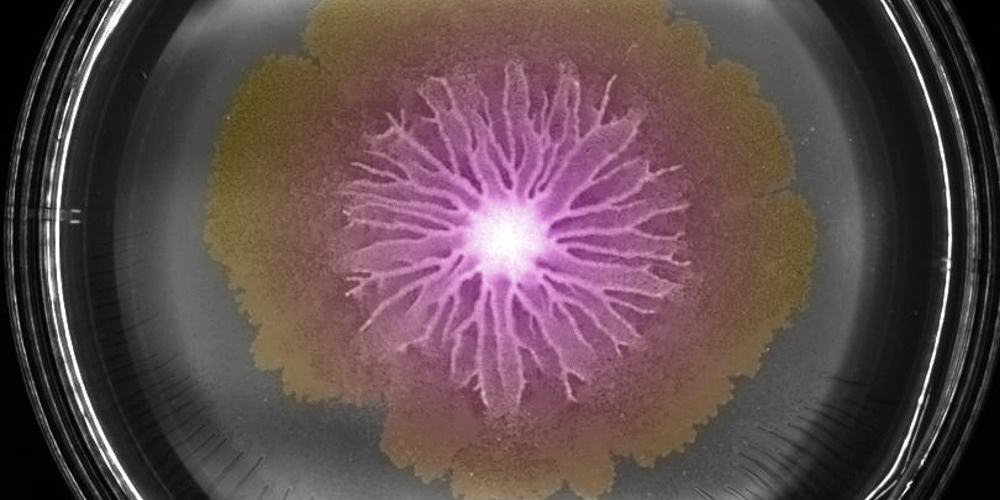 Swarm of Bacillus subtilis bacteria on an agar plate. Image Credit: Biozentrum, University of Basel.
Swarm of Bacillus subtilis bacteria on an agar plate. Image Credit: Biozentrum, University of Basel.
Bacteria usually live in communities in natural settings. They produce biofilms like dental plaque or collectively colonize our gut, sometimes referred to as the gut microbiome. For individual bacteria, living in groups has several benefits. They can conquer new lands, withstand harsh weather conditions, and gain from one another more effectively.
Analyzing microbial communities in space and time
Bacteria create elaborate three-dimensional structures during the extremely complicated process of bacterial community growth. The group under the direction of Professor Knut Drescher from the Biozentrum of the University of Basel has examined the evolution of bacterial swarm communities in great depth in their most recent work, which was published in Nature Microbiology.
They made a methodological breakthrough that allowed them to visualize the behavior of individual cells in microbial communities in both space and time, as well as assess gene expression simultaneously.
Bacteria provide resources for future generations
We used Bacillus subtilis as a model organism. This ubiquitous bacterium is also found in our intestinal flora. We have revealed that these bacteria, which live in communities, cooperate and interact with each other across generations.”
Knut Drescher, Study Head and Professor, University of Basel
Additionally, they discovered that within a bacterial swarm, there are distinct subpopulations that generate and utilize distinct metabolites. Subpopulations that form later in the development of swarms eat some of the metabolites secreted by one subpopulation.
Distribution of tasks within the community
Modern adaptive microscopy, metabolite analysis, gene expression analysis, and robotic sampling were all combined by the researchers. With this novel method, the researchers have identified the metabolites released by the bacteria and have been able to concurrently study gene expression and bacterial behavior at well-defined times and locations.
Thus, the swarm front, intermediate region, and swarm center might be considered the three main regions of the bacterial swarm. All three regions, however, show progressive changes.
Depending on the region, the bacteria differ in appearance, characteristics and behavior. While they are mostly motile at the edges, the bacteria in the center form long non-motile threads, resulting in a 3D biofilm. One reason is the varying availability of space and resources. The spatial distribution of bacteria with distinct behavior enables the community to expand but also to hide in a protective biofilm.”
Hannah Jeckel, Study First Author and Postdoctoral Scholar Research Associate, Biology and Biological Engineering, California Institute of Technology
Complex dynamics within bacterial communities
This study uncovers cooperative relationships among individual bacteria that are advantageous to the group, as well as the intricacy and dynamism inside bacterial communities. Hence, the formation and growth of microbial communities are significantly influenced by geographical and temporal circumstances.
The creation of a novel method that allowed the researchers to gather complete spatiotemporal data on a multicellular process at a resolution never previously possible in any other biological system is a significant accomplishment of this work.
Source:
Journal reference:
Jeckel, H., et al. (2023). Simultaneous spatiotemporal transcriptomics and microscopy of Bacillus subtilis swarm development reveal cooperation across generations. Nature Microbiology. doi.org/10.1038/s41564-023-01518-4.