Analytical techniques like atomic force microscopy (AFM) enable researchers to observe the functional dynamics of molecular structures like proteins directly. However, sample placement, which must be kept immobile to obtain accurate AFM results, cannot be directly controlled during investigations. By employing a computational-based approach, researchers from Kanazawa University have introduced a new method for predicting protein placement on AFM substrates.
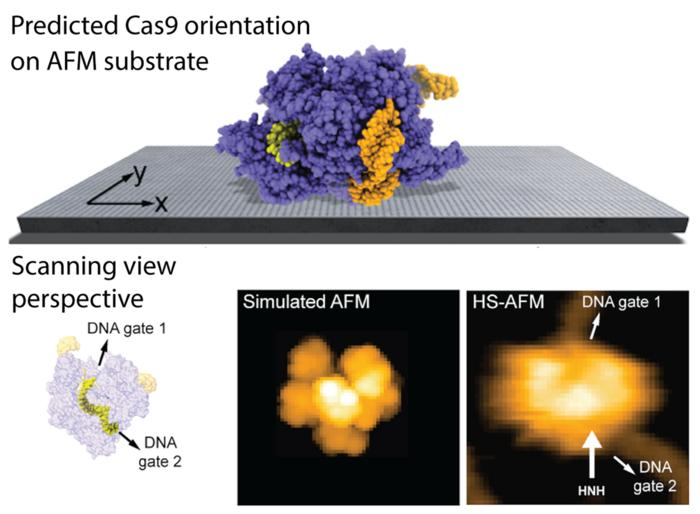 The predicted orientation of the Cas9 protein on the AFM substrate is shown, and a simulated AFM image calculated in the scanning view perspective shows remarkable agreement with previous HS-AFM imaging. HS-AFM can reliably visualize functional relative motions of target DNA and Cas9 because the entry and exit gates of the DNA strand, which is located in a tunnel within the Cas9 structure, are not blocked by contacts with the APTES-mica substrate. Image Credit: 2023 Amyot, Nakamoto, Kodera and Flechsig.
The predicted orientation of the Cas9 protein on the AFM substrate is shown, and a simulated AFM image calculated in the scanning view perspective shows remarkable agreement with previous HS-AFM imaging. HS-AFM can reliably visualize functional relative motions of target DNA and Cas9 because the entry and exit gates of the DNA strand, which is located in a tunnel within the Cas9 structure, are not blocked by contacts with the APTES-mica substrate. Image Credit: 2023 Amyot, Nakamoto, Kodera and Flechsig.
The method, as detailed in Frontiers in Molecular Biosciences and lead by Romain Amyot, Noriyuki Kodera, and Holger Flechsig, harnesses electrostatic interactions and has specific applications to high-speed AFM (HS-AFM) observations of highly dynamic protein samples like Cas9 endonucleases.
The Intricacies of AFM and HS-AFM Imaging
AFM and HS-AFM offer scientists the power to observe biomolecular structures and their functional dynamics directly under near-physiological conditions. Typically, biological samples of interest are placed on an AFM substrate, or supporting surface, and are scanned by a probe tip to determine their molecular shape. Any dynamical changes that occur are also detected.
However, when loading these samples, a delicate balance must be maintained: the sample must be immobilized on the AFM's supporting surface with as little interference to its function in a biological system as possible.
Substrate Modelling and Predicting Protein Placement
The new model, developed within the BioAFMviewer software, utilizes an electrostatic interaction model to predict how proteins interact with various AFM substrates like mica, APTES-mica, and lipid bilayers under different buffer conditions. This model takes into account the Debye-Hückel potential, considering long-range interactions as the primary factor influencing biomolecular placement on the substrate.
Using the software, the substrates are modeled in detail, accounting for their specific charge properties and surface modifications, which are critical for successful AFM imaging.
The researchers synthetically generated a variety of biomolecular orientations on the substrate from a limited set of manually annotated images. Doing so allowed for predictions of the most favorable placements based on evaluating interaction energies and the stability of imaging under tip scanning.
Notably, the team demonstrated the validity of their approach with applications to HS-AFM imaging of key proteins like the Cas9 endonuclease, offering insights into the stability of observed biomolecular interactions and confirming resolution-limited imaging results using atomistic-level information.
In the case of the Cas9 endonuclease, the research findings are able to visualize functional relative motions of target stands of DNA and Cas9, capturing cleavage events at a single molecular scale, as shown in the figure above.
The model is also able to explain how the stability of sample-substrate complexes can be influenced by buffer conditions. .
Applications and Implications for Future Research
Using this computational-based technique, researchers are able to use existing structural data of biomolecules to predict sample placement on AFM substrates even before conducting an actual experiment. It also proves useful for post-experimental analysis of AFM imaging data.
The implications of this work extend beyond methodological advancement; by providing a more nuanced understanding of how proteins interact with substrates, more accurate and detailed observations of biomolecular dynamics can be obtained.
Such insight is essential for advancing our understanding of biological processes at the nanoscale and stands to be a valuable tool for the broader Bio-AFM community.
Further Reading
Source:
Journal reference:
Amyot, R., et al (2023). Predicting the placement of biomolecular structures on AFM substrates based on electrostatic interactions, Frontiers in Molecular Biosciences. 10. DOI: https://doi.org/10.3389/fmolb.2023.1264161