The revolution in plant species characterization, brought about by advancements in whole-genome sequencing, has yielded a vast pool of genotypic data available for analysis.
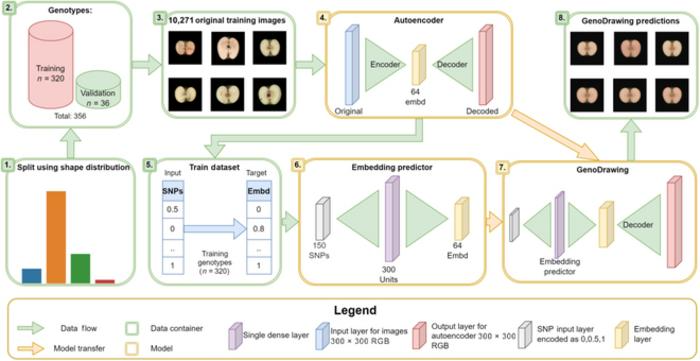 Graphical abstract of the general approach. Image Credit: Plant Phenomics.
Graphical abstract of the general approach. Image Credit: Plant Phenomics.
Integrating genomic selection with neural networks, particularly deep learning and autoencoders, presents a promising approach for predicting intricate traits from this data. Despite its success in applications such as plant phenotyping, accurately translating visual information from images into quantifiable data for genomic studies still poses challenges.
The study was published on November 2023, in the journal Plant Phenomics.
The research introduces a novel approach that employs an autoencoder network and embedding predictor to condense apple images into 64 dimensions and predict fruit shapes based on molecular data (SNPs).
The methodology, termed GenoDrawing, entails training the autoencoder using an extensive dataset of apple images. Subsequently, the generated embeddings, in conjunction with SNP data, are utilized for the prediction and reconstruction of apple shapes.
The results demonstrated that targeted SNPs (tSNPs) consistently outperformed randomly selected SNPs (rSNPs) in predicting image embeddings, leading to more precise predictions of fruit shapes.
The top-performing models utilizing targeted SNPs (tSNPs) exhibited lower mean absolute errors (MAEs) and generated distributions that closely resembled the original data, in contrast to models employing randomly selected SNPs (rSNPs). Moreover, the tSNP-based version demonstrated its efficacy by predicting a broader range of fruit shapes, showcasing its effectiveness in capturing the diversity of apple phenotypes.

Image Credit: Nataly Studio/Shutterstock.com
Nevertheless, the study uncovered certain limitations, such as the model's challenge in accurately capturing specific fruit features and the impact of environmental factors on apple phenotypes. Despite these hurdles, the approach signifies a noteworthy stride in genomic prediction, showcasing the potential of integrating image analysis with molecular data to comprehend intricate crop traits.
To summarize, the results indicate the significance of choosing pertinent SNPs for precise predictions, with GenoDrawing demonstrating efficacy in learning to predict fruit shapes when provided with suitable markers. This research establishes a foundation for future studies seeking to refine the accuracy and applicability of genomic prediction models by integrating image data and enhancing SNP selection strategies.
Source:
Journal reference:
Jurado-Ruiz, F., et al. (2023). GenoDrawing: An autoencoder framework for image prediction from SNP markers. Plant Phenomics. doi.org/10.34133/plantphenomics.0113.