Reviewed by Danielle Ellis, B.Sc.Jan 30 2024
The mapping of genetic blueprints for 51 species, such as sharks, turtles, kangaroos, cats, penguins, and dolphins, has advanced the knowledge of evolution and the relationships between animals and people.
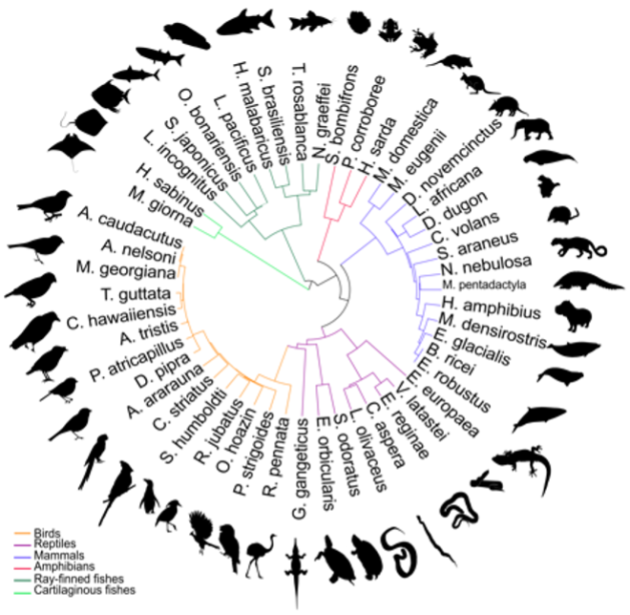 Researchers selected these 51 animals for gene sequencing, deepening our understanding of how they are related to each other. Image Credit: Delphine Larivière, Penn State University
Researchers selected these 51 animals for gene sequencing, deepening our understanding of how they are related to each other. Image Credit: Delphine Larivière, Penn State University
Being able to access that genetic information will have huge implications for understanding human health and evolution, and a lot of work on drug compounds starts in mice and other animal models, so understanding their genomes and the genomes of other animals directly benefits us."
Michael Schatz, Study Lead Author and Bloomberg Distinguished Professor, Department of Biology and Oncology, Johns Hopkins University
Collaborating with the Vertebrate Genomes Project, the team undertook the sequencing of genomes from 51 vertebrate species, focusing on those serving as valuable models for comprehending human evolution.
Employing innovative algorithms and computer software, the researchers significantly reduced the sequencing time from months, or even decades in the case of the human genome, to a mere matter of days.
The journal Nature Biotechnology published the findings.

Image Credit: natrot/Shutterstock.com
Almost all the genes from a common ancestor who lived about 200 million years ago and 50% to 99% of the DNA are shared by mammals, a subset of vertebrates that includes humans, dogs, cats, monkeys, and mice.
Researchers can begin to determine when and where DNA sequences diverged and the consequences of those changes for humans by comparing the entire genomes of different species. However, due to the scarcity and poor quality of vertebrate genomes, which have mostly concentrated on a small number of important species, researchers claim that their work has been hampered.
Since the genomes of vertebrates contain billions of characters, no gene sequencing method can read them all at once. Scientists are forced to rely on methods that divide the genome into more manageable, readable chunks.
After that, the genome of the zebra finch, a songbird whose genome had previously been sequenced to examine brain development, was mapped by researchers as a test of their method. The genome could now be reassembled much more accurately and completely thanks to the new technology.
The open-source software can be accessed online through Galaxy, a web-based platform hosted at Penn State and Johns Hopkins that supports 500,000 scientists and educators globally and provides free scientific software to the public., computer programs take those segments and figure out how they go together, much like jigsaw puzzle pieces.
However, conventional technology was unable to solve the puzzle.
Have you ever done a massive jigsaw puzzle where at some point all that is left is blue sky, and you do not think you will ever be able to fit the right pieces together? The old software would give up on these hard parts of the genome. That is the problem with genome assembly, and our new program, using the latest sequencing data and the latest assembly algorithms, knows how to work through those parts to get a more complete picture."
Michael Schatz, Study Lead Author and Bloomberg Distinguished Professor, Department of Biology and Oncology, Johns Hopkins University
The genome of the zebra finch, a songbird whose genome had previously been sequenced to examine brain development, was mapped by researchers as a test of their method. The genome could now be reassembled much more accurately and completely thanks to the new technology.
The open-source software can be accessed online through Galaxy, a web-based platform hosted at Penn State and Johns Hopkins that supports 500,000 scientists and educators globally and provides free scientific software to the public.
In the past, only a handful of elite research groups would have had access to the resources needed to assemble these genomes. Now, anyone on the planet with access to the internet can visit the website and, with a few clicks of the button, run multiple scientific tools."
Alex Ostrovsky, Software Engineer, Galaxy Team, Johns Hopkins University
He is also responsible for making the tools easy to use for noncoders.
To sequence the genomes of at least one species from each of the 275 vertebrate orders, the researchers will keep collaborating with the Vertebrate Genomes Project.
Schatz said, "In some ways, we are building an evolutionary time machine, and we can trace how vertebrates evolved over time and eventually gave rise to genes and sequences that are uniquely found in humans. Having the genes of our evolutionary cousins mapped out will help us better understand ourselves."
Source:
Journal reference:
Larivière, D., et.al., (2024). Scalable, accessible, and reproducible reference genome assembly and evaluation in Galaxy. Nature Biotechnology. doi.org/10.1038/s41587-023-02100-3