In 2003, we saw the human genome fully sequenced for the first time. Since that landmark accomplishment, there have been numerous technological advancements in sequencing technology, not just in terms of DNA but also within the context of protein interactions and DNA - known as Chromatin immunoprecipitation sequencing (ChIP-seq).
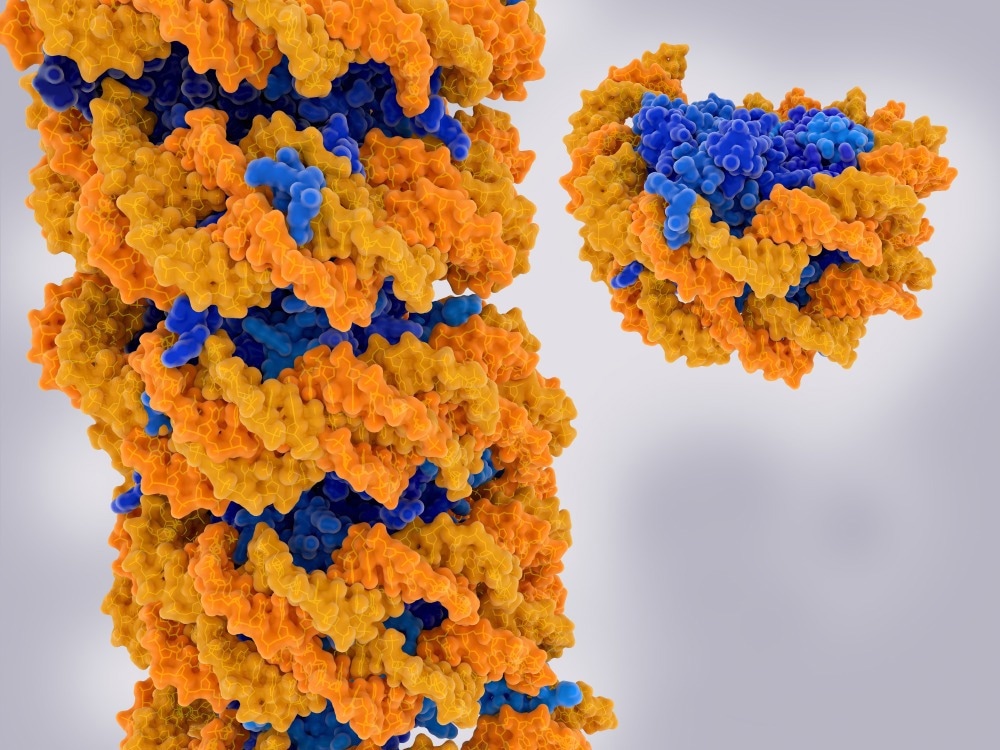 Chromatin strand and isolated nucleosome. Chromatin is a complex of DNA (yellow) and proteins (blue). Nucleosomes form the fundamental repeating units. Image Credit: Juan Gaertner/Shutterstock.com
Chromatin strand and isolated nucleosome. Chromatin is a complex of DNA (yellow) and proteins (blue). Nucleosomes form the fundamental repeating units. Image Credit: Juan Gaertner/Shutterstock.com
Chromatin comprises proteins (such as histones and transcription factors) and DNA, with this complex located within the nucleus of a cell. ChIP-seq involves working with the chromatin structure in order to isolate and study the DNA-protein interactions present, thus generating valuable information, such as the target protein binding site upon the DNA, consequently furthering our understanding of epigenetics and gene regulation.
The initial Chromatin Immunoprecipitation (ChIP) technique was pioneered by Gilmour and Lis presented in their 1984 paper during their investigation of the interaction between RNA polymerase II and genes in Escherichia coli and Drosophila. In their early work, UV irradiation was employed to create covalent bonds between proteins and nearby DNA within living cells.
Later on, however, Solomon and Varshavsky replaced the UV cross-linking method with the formaldehyde cross-linking approach, which is still used today. ChIP-seq, as we know it, was developed by Robertson and colleagues in 2007 and used to pinpoint DNA sequences in mammals bound by transcription factors inside living cells.
Technological advancements in ChIP-seq have revolutionized the study of chromatin biology and gene regulation. The advent of Next-Generation Sequencing (NGS) platforms, notably Illumina, has significantly increased throughput and reduced costs. Single-cell ChIP-seq allows researchers to explore cellular heterogeneity. Further innovations such as ChIP-exo and ChIP-nexus have enhanced resolution and accuracy in mapping protein-DNA interactions.
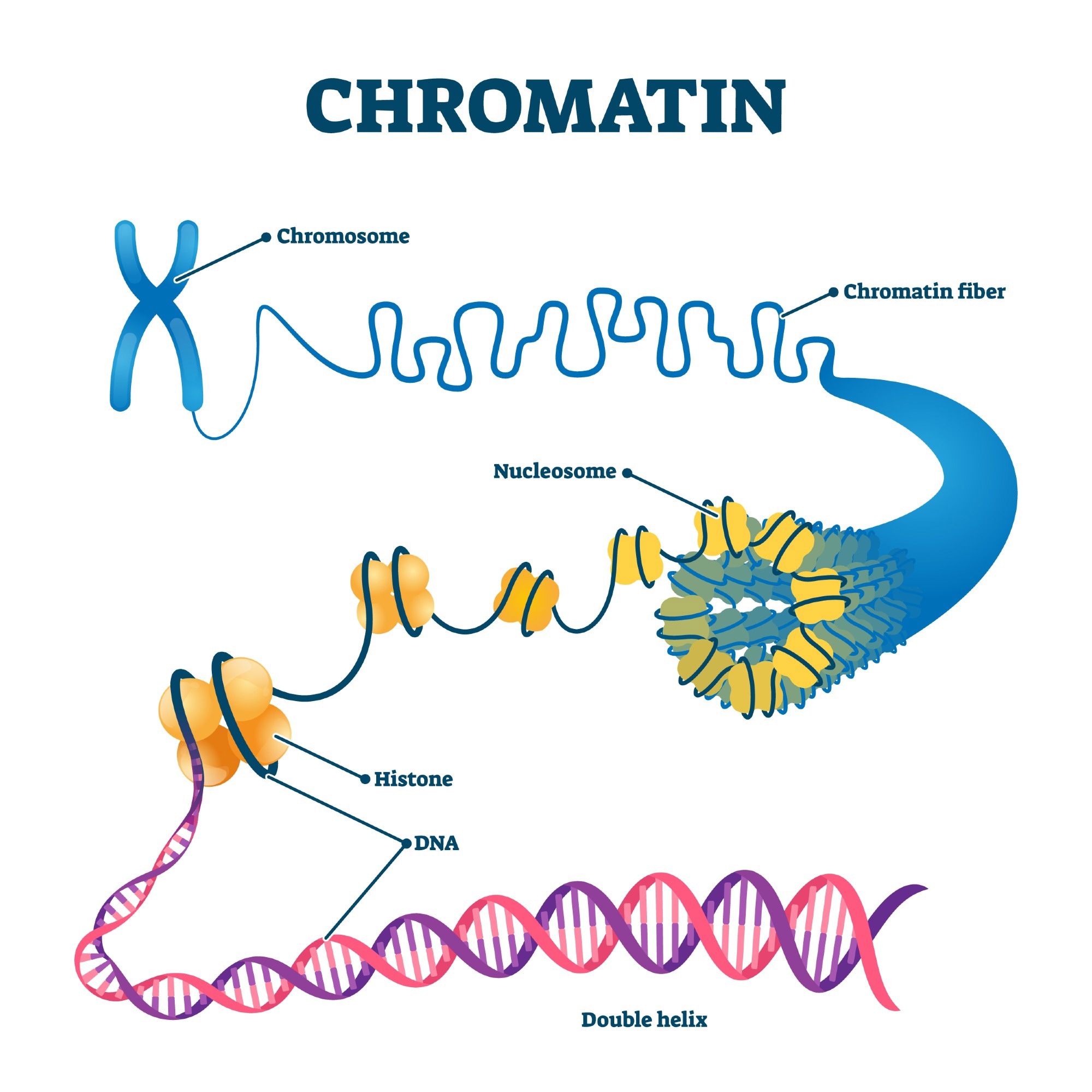 Chromation biological diagram vector illustration. Close-up with nucleosome, histone and DNA double helix. Science educational information. Chromosome structure elements graphic example model. Image Credit: VectorMine/Shutterstock.com
Chromation biological diagram vector illustration. Close-up with nucleosome, histone and DNA double helix. Science educational information. Chromosome structure elements graphic example model. Image Credit: VectorMine/Shutterstock.com
Understanding Epigenetics and Gene Regulation
Epigenetics involves heritable changes in gene function beyond the DNA sequence, crucial for regulating gene expression, whereby mechanisms such as DNA methylation, histone modifications, and non-coding RNAs impact chromatin structure and thus have vital roles in cellular diversity, development, and disease. Such epigenetic modifications dynamically control gene expression, cellular differentiation, and responses to the environment. Hence, with its dysregulation, diseases may arise, which therefore underlies the importance of understanding epigenetics to uncover gene regulation complexities.
ChIP-seq allows the mapping of protein-DNA interactions associated with chromatin modifications, thereby allowing for the identification of genomic locations enriched with specific histone modifications or DNA-binding proteins, thus offering a genome-wide view of epigenetic modifications, as well as transcriptional regulation, contributing significantly to our understanding of epigenetic gene regulation.
The Process of ChIP-Seq
In ChIP-seq experiments, a series of carefully orchestrated steps are undertaken to unravel the intricate interactions between proteins and DNA. The process begins with crosslinking, where cells are treated to fix protein-DNA interactions, which is then followed by cell lysis in order to induce chromatin fragmentation - liberating chromatin into smaller fragments.
Immunoprecipitation (IP) comes next, where antibodies selectively bind to the target protein or modifications, forming distinct complexes. Subsequent washing removes unbound DNA, leaving only fragments bound to the target protein, followed by the reversal of crosslinks, thereby effectively separating proteins from DNA.
To ensure high-quality samples, DNA is purified before undergoing library preparation. During this step, DNA is fashioned into a sequencing library, incorporating sequencing adapters for later analysis. High-throughput sequencing, often facilitated by platforms like Illumina, follows, generating vast amounts of data.
The bioinformatics analysis phase involves mapping reads, identifying peaks, and annotating enriched regions. The culmination of this process is interpretation, where results are analyzed to unveil valuable insights into protein-DNA interactions and the broader functionality of chromatin.
Role of Antibodies in Targeting Proteins or Modifications
Antibodies are pivotal in ChIP-seq through their selective isolation of protein-DNA complexes. Chosen for their specificity, antibodies target proteins or modifications like transcription factors or histones. Acting as molecular tools, antibodies allow researchers to pull down specific targets, offering a focused view of genomic regions involved in gene regulation. Antibody specificity is critical for accurate ChIP-seq results, determining the precision with which protein-DNA interactions are identified within the complex chromatin structure.
Applications of ChIP-sequencing in Research
It is thus that Chip-seq has been adopted in research, assisting in studying molecular phenomena of gene expression and hence the genome-wide map of transcription factor binding sites upon DNA as well as epigenetic signatures, i.e., susceptibility to DNA methylation. As such, it has been utilized to study the regulation of genes and epigenetic mechanisms, especially in the diseased context.
For the dog lovers out there, you will be pleased to know that ChIP-seq has not only been employed in research regarding humans, whereby ChIP-seq, alongside whole genome and transcriptome data analysis, was utilized to identify mutations within the canine genetic loci on chromosomes 5 (CFA5) that are associated with histiocytic sarcoma in flat-coated retrievers - histiocytic sarcoma being an aggressive form of cancer affecting immune cells, although rare in humans.
Published in the PLOS Genetics journal, 2021, researchers using a genome-wide association survey identified two loci within the canine genome, that being CFA5 and CFA19, associated with histiocytic sarcoma. From there, the researchers then utilized whole genome, transcriptome, and CHiP-seq data to identify mutations near the tumor-suppressor gene PIK3R6 within the canine genetic locus of CFA5.
Also, they found increased TNFAIP6 expression at the canine genetic locus on chromosome 19 (CFA19) via other research tools (RNAseq). The combined impact of such genetic factors at both CFA5 and CFA19 accounted for a substantial 35% of the risk of histiocytic sarcoma in flat-coated retrievers. Beyond canine health, such findings offer translational insights for potential diagnostics and therapeutics in human oncology.
Conclusion
In summary, ChIP-sequencing (ChIP-seq) plays an important role in advancing our understanding of epigenetic modifications and gene regulation, proving to be significant not only for scientific research within the human sphere but also for expanding its use in veterinary research, illustrating its universal applications.
By mapping protein-DNA interactions associated with chromatin modifications, ChIP-seq provides valuable insights and elucidates the complexity that is epigenetics and gene regulation, thereby shedding light on how factors like DNA methylation, histone modifications, and non-coding RNAs impact cellular processes and, most importantly, within the context of disease as to pave the way for novel therapeutic avenues for clinical application.
References
- Lloyd SM, Bao X. Pinpointing the Genomic Localizations of Chromatin‐Associated Proteins: The Yesterday, Today, and Tomorrow of ChIP‐seq. Current Protocols in Cell Biology. 2019 Jun 3;84(1).
- Gilmour, D. S. and Lis, J. T. (1984) Proceedings of the National Academy of Sciences, 81(14), pp. 4275–4279. doi: 10.1073/pnas.81.14.4275.
- Solomon, M. J. and Varshavsky, A. (1985) Proceedings of the National Academy of Sciences, 82(19), pp. 6470–6474. doi: 10.1073/pnas.82.19.6470.
- Robertson, G., Hirst, M., Bainbridge, M. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4, 651–657 (2007). https://doi.org/10.1038/nmeth1068
- Mundade R, Ozer HG, Wei H, Prabhu L, Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle. 2014 Sep 17;13(18):2847–52.
- Rhee HS, Pugh BF. ChIP‐exo Method for Identifying Genomic Location of DNA‐Binding Proteins with Near‐Single‐Nucleotide Accuracy. Current Protocols in Molecular Biology. 2012 Oct;100(1).
- He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nature Biotechnology. 2015 Mar 9;33(4):395–401.
- Thermo Fisher Scientific. Step-by-Step Guide to Successful ChIP Assays. [Internet]. Thermo Fisher Scientific; c2016 [cited 2024 Feb 4]. Available from: https://assets.thermofisher.com/TFS-Assets/LSG/Application-Notes/Step-by-Step-Guide-to-Successful-ChIP-Assays.pdf
- Asp P. The chromatin immunoprecipitation (ChIP) assay and ChIP-qPCR. Elsevier eBooks. 2020 Jan 1;281–96.
- Angelini C, Costa V. Understanding gene regulatory mechanisms by integrating ChIP-seq and RNA-seq data: statistical solutions to biological problems. Frontiers in Cell and Developmental Biology. 2014 Sep 17;2.
- Mancarella D, Plass C. Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation. Genome Medicine. 2021 Feb 10;13(1).
- Park PJ. ChIP–seq: advantages and challenges of a maturing technology. Nature Reviews Genetics [Internet]. 2009 Sep 8;10(10):669–80. Available from: https://www.nature.com/articles/nrg2641
- Evans J, Parker HG, Rutteman GR, Plassais J, Guy, Harris AC, et al. Multi-omics approach identifies germline regulatory variants associated with hematopoietic malignancies in retriever dog breeds. PLOS Genetics. 2021 May 13;17(5):e1009543–3.
Further Reading