Top-down and bottom-up approaches and models can be used for a range of different fields, including psychology, information processing, nanotechnology, engineering, and physiological processing. Within systems biology, using top-down analysis can enable correlations to be found in genes and proteins within a large data set – this can be useful for finding connections to build upon for the creation of an experiment. However, this can be difficult to do in reality.
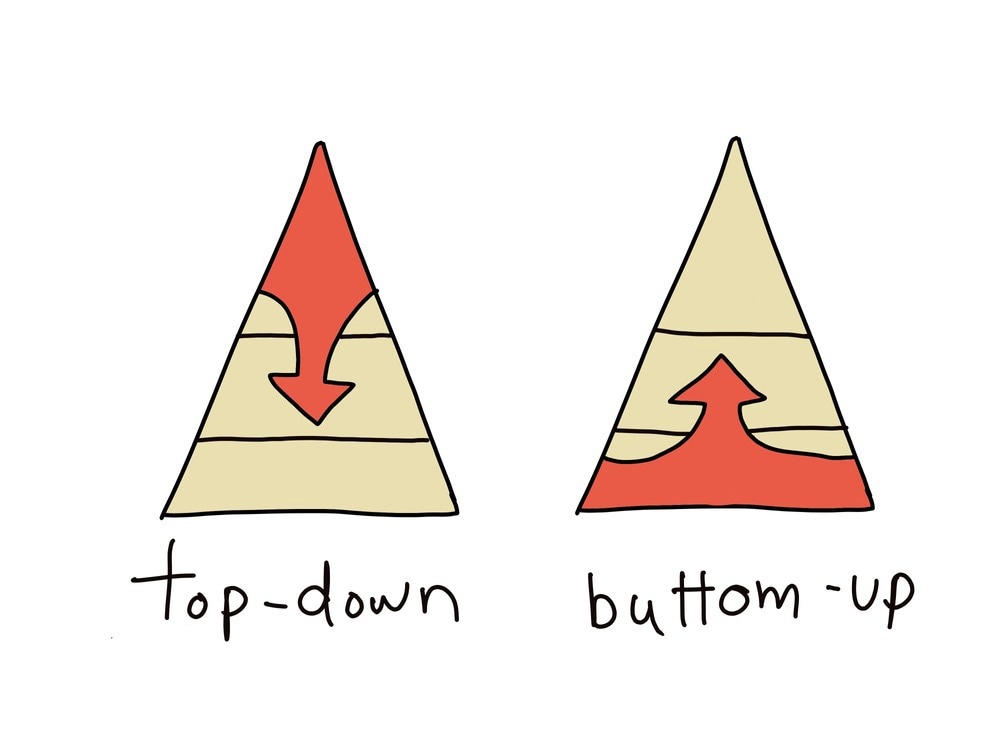
Image Credit: DaViDa S/Shutterstock.com
A bottom-up approach may be more useful as this model includes details that enable computational simulations that can be used as the basis of wet lab experimentation.
The metabolic behavior of a cell can be approached with either of the two models, with the top-down approach including data processing from a higher analytic ‘omics’ level, such as proteomics or genomics, to pathways and individual genes of certain organisms. The bottom-down approach, however, can include developing automated tools and using mathematical models.
Researchers, Oltvai and Barabasi, have modeled both top-down and bottom-up approaches within a pyramid to depict two different levels consisting of ‘organism specificity’ and ‘universality’. These can be used to show how a cell can be approached equally within both approaches, from bottom to top, referring to ‘universality’, which include the creation of scale-free networks and modules from molecules, and top to bottom, which reflects ‘organism specificity’, which includes moving from scale-free networks to specific organisms.
Using Bottom-Up Approaches
Bottom-up approaches consist of constructing models to manufacture an experimentational plan through simulations under a range of physiological conditions.
This approach can combine organism-specific information within a complete genome-scale model to aid the assessment and analysis of biological interactions that occur inside a living system. This can enable the building of genome-scale mathematical models using four key steps, including (i) draft reconstruction, (ii) manual curation, (iii) conversion of curated models into a mathematical format, and (iv) validation of the models through literature reviews, biochemical assays, and data on ‘omics’.
Draft reconstruction consists of collecting data from various online resource databases, including genomics, biochemical, metabolic, and organism-specific information.
Manual curation can include human involvement and software assistance to add missing data and remove irrelevant information on organism-specific genomes. Once a curated draft is complete, it can be transformed into a mathematical form/language to create a model and perform simulations. This model finally undergoes validation, where it is refined and assessed for inconsistencies using objective functions and various gap-filling algorithms.
This process outputs a final accurate genome-scale computational model for system biology, which can be used for possible wet lab experimentation for researchers.
Using Top-down Approaches
The top-down approach is derived from experimental information that can be used to reconstruct metabolic models, which may be useful for understanding biological behavior and the interactions of organisms at a basic level. This can be garnered through ‘omics’ data that can be provided through standard genome methods, including DNA microarrays and RNA sequencing.
Researchers such as Van Dien and Schilling have stated the flow of data within this approach is from the transcriptome and proteome, including RNA and proteins, to metabolic pathways.
Image Credit: Andre Nantel/Shutterstock.com
Top-down Stages
The top-down approach can have five different stages, which include (i) sample collection and experimentation, (ii) high-throughput genomics using DNA microarrays and RNA sequencing, (iii) statistical analysis, (iv) applying bioinformatics, and (v) data interpretation and discovery.
This can be summarized with a sample collected from a particular treatment or stimuli given to animals, which can demonstrate the impact on the animal’s physiology. Using high-throughput genomics, such as a DNA microarray platform, can determine mRNA expression levels within particular cells or tissues, which then undergoes statistical analysis.
Statistical probability values or p-values can be produced when determining differentially expressed genes and can be useful when experiments contain multiple comparisons to aid researchers in understanding the significance of their results. Additionally, bioinformatics can aid in annotating genes and pathways to assess the impact of various stimuli or treatments on differentially expressed genes.
Ultimately, the resulting pathway can be analyzed using scientific articles and organism-specific databases, which can be used when analyzing the whole genome of an organism. This can be beneficial for researchers attempting to understand the molecular behavior of genes and proteins placed under various environmental conditions and physiological states.
The use of top-down models can be useful for researchers that have found a pattern or molecular behavior and are investigating the underlying mechanism.
Future Outlook
Both bottom-up and top-down approaches can be used within genomic research to investigate and understand complex biological mechanisms still obscure within the scientific community. Utilizing these methods can either ensure computational models are created for accurate experimental plans or for the investigation of samples to understand genetic behavior at a molecular level.
These approaches can enhance knowledge within science and advance fields such as medicine by producing drugs for biological targets to prevent human disease.
Sources:
- Gregorich Z, Ge Y. Top-down proteomics in health and disease: Challenges and opportunities. Proteomics. 2014;14(10):1195-1210. doi:10.1002/pmic.201300432
- Poirel C, Rodrigues R, Chen K, Tyson J, Murali T. Top-Down Network Analysis to Drive Bottom-Up Modeling of Physiological Processes. Journal of Computational Biology. 2013;20(5):409-418. doi:10.1089/cmb.2012.0274
- Shahzad K, J. Loor J. Application of Top-Down and Bottom-up Systems Approaches in Ruminant Physiology and Metabolism. Curr Genomics. 2012;13(5):379-394. doi:10.2174/138920212801619269
Last Updated: Nov 9, 2022