Over the last few years, genomics research has been fundamental for developing diverse technologies, from agriculture to medicine. In agriculture, genomics has identified significant genes of agronomic importance and aided the production of higher-yield crops. In medicine, genomics has been critical for designing effective and safe vaccines, tracing pathogenic strains in epidemiology, mapping disease loci in whole-genome association studies, assessing gene expression patterns in cancer cells, etc.

Image Credit: Yurchanka Siarhei/Shutterstock.com
Genomics has become one of the most important disciplines in life sciences. It is almost impossible to imagine a future without genomics applications in our daily life. Genomic technologies have shed light on the functioning and organization of hereditary material and the diversity in the tree of life. This discipline represents a rosetta stone for the generation of new methods in disease risk analysis, high-scale production of human hormones, molecular breeding, pest management strategies, etc.
What does genomics mean?
The genome is the complete genetic material (DNA) within a living cell. Genomics is a discipline aimed at studying the organization and structure of the genome within the cell nucleus. The linear order of nucleotides in discrete DNA molecules (e.g., linear chromosomes) is key for understanding many aspects of biology, such as, for example, the evolutionary history of a lineage from its root ancestor, developmental signaling pathways associated with the formation of tissues, diseases states, etc.
Current technologies in genomics research
Functional genomics is a major branch of genomics research that deals with studying the genome at all' omic levels. Functional genomics enables the combination of distinct sources of omics data, including gene expression data (transcriptomics), epigenetic modifications (epigenomics), and protein abundance (proteomics), thus understanding how a genome works at different molecular levels to shape a phenotype. Some of the latest and most widely extended advancements in functional genomics research came from technologies such as RNA interference (RNAi), genome-wide mutagenesis, and genome engineering. Moreover, the emergence of molecular systems to precisely edit and modify genomes, especially the arrival of the programmable CRISPR-Cas9 genome editing system, has also provided a wealth of opportunities in genomics research.
The RNA interference (RNAi) pathway is an evolutionarily conserved process that uses double-stranded regulatory RNAs (dsRNAs) to silence target genes by downregulating their expression. The RNAi pathway was observed as a naturally occurring mechanism in the model organism Caenorhabditis elegant by Andrew Fire and Craig Mello (1998), a discovery that led them to win Nobel Prize in Physiology in 2006. In the laboratory, dsRNAs can be easily designed to target messenger RNA (mRNA) sequences to repress gene expression. The regulatory RNAi pathway has already been exploited in many fields (e.g., the study of disease-causing genes) by their inhibition in a sequence-specific manner.
Genome-wide mutagenesis is another method widely used in genomics research to identify genes whose expression is associated with a phenotype of interest. This technique generates random mutations scattered at multiple loci across the genome. Genome-wide mutagenesis has been a powerful tool used in reverse genetics to generate multiple loss-of-function mutations at different loci involved in expressing a phenotypic trait.
Moreover, the CRISPR-Cas9 system is an RNA-programmable genome editing system widely used to edit genomes in a targeted and cost-effective manner. Since its development in 2013, the CRISPR-Cas9 system has sparked a revolution in genomics. Multiplexed CRISPR technologies have also enabled the functional identification of genes in virtually any organism via selective targeting of specific genomic loci using single-guide RNA (sgRNA) sequences.
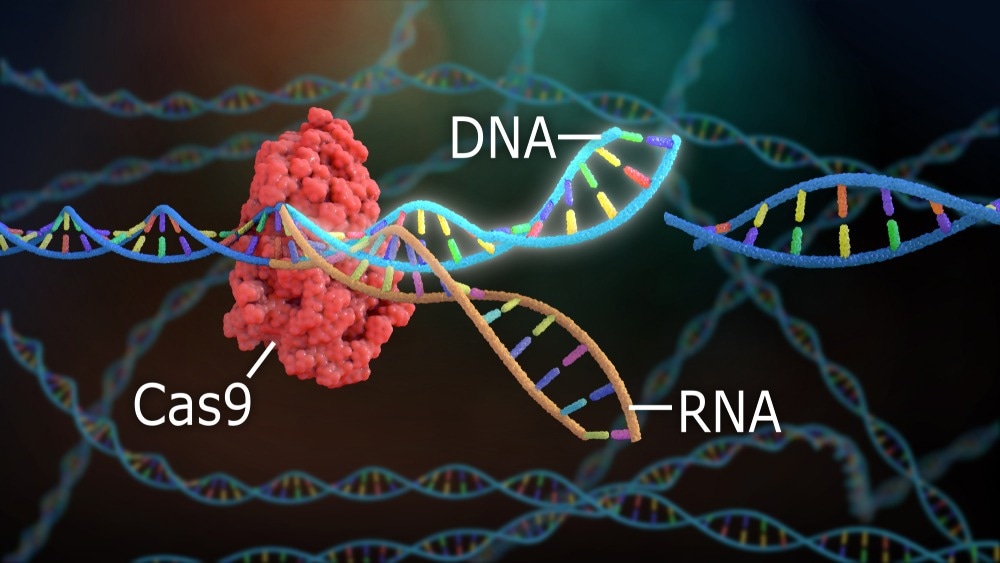
Image Credit: Nathan Devery/Shutterstock.com
There is a bright tomorrow in genomics
The combination of faster and more effective bioinformatic algorithms for analyzing genomic data and new cost-effective next-generation sequencing platforms for obtaining massive volumes of sequence data in real time allows for predicting a promising future in genomics research.
Recent advancements in molecular biology will also surely expand the field of applications in genomics research. For instance, the development of RNA activation (RNAa) strategies capable of activating target genes by small activating RNAs (saRNAs) can be integrated with RNAi procedures to shape whole genome expression in a programmable RNA-guided manner accurately. Finally, the emergence of a myriad of CRISPR-based technologies is already achieving the design of new applications in genomics, such as site-specific epigenetic modifications, transcriptional targeting, cell-site imagining, etc.
Sources:
- Ahmed, A. E., Allen, J. M., Bhat, T., Burra, P., Fliege, C. E., Hart, S. N., ... & Mainzer, L. S. (2021). Design considerations for workflow management systems use in production genomics research and the clinic. Scientific reports, 11(1), 1-18.
- Hindorff, L. A., Bonham, V. L., Brody, L. C., Ginoza, M. E., Hutter, C. M., Manolio, T. A., & Green, E. D. (2018). Prioritizing diversity in human genomics research. Nature Reviews Genetics, 19(3), 175-185.
- Lu, Y., Ye, X., Guo, R., Huang, J., Wang, W., Tang, J., ... & Qian, Y. (2017). Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Molecular Plant, 10(9), 1242-1245.
- Novina, Carl D., and Phillip A. Sharp. "The rnai revolution." Nature 430.6996 (2004): 161-164.
- Nunn, Jack S., et al. "Public involvement in global genomics research: a scoping review." Frontiers in public health 7 (2019): 79.
- Wang, Fangyuan, and Lei S. Qi. "Applications of CRISPR genome engineering in cell biology." Trends in Cell Biology 26.11 (2016): 875-888.
Further Reading
Last Updated: Oct 26, 2022