Genetic drift is one of the four main factors associated with changes in the gene pool over time. Other factors that lead to genetic changes are mutation, natural selection, and gene flow. This article focuses on genetic drift and its consequences.

Image Credit: adike/Shutterstock.com
Genetic Drift vs. Natural Selection
It is important to understand the difference between genetic drift and natural selection. The theory of natural selection states that a population harbors individuals with varied alleles. For instance, some individuals possess favorable alleles for the environment in which they live that aid their survival and reproduction. Other individuals bearing unfavorable alleles for their environment are responsible for detrimental effects that ultimately result in their death before reproduction. After many successive generations, the selection pressure leads to a change in the genetic pool and traits within a population.
In contrast, genetic drift describes the effect of chance on a population without any selection pressure. This theory entails a shift of alleles within a population due to a chance event that causes random samples within a population to reproduce or not.
Genetic Drift and Its Consequence
Genetic drift has been defined as a random change in allele frequencies between generations in a particular population due to an unplanned sampling event. It occurs due to the presence of a differential survival rate or fecundity that is distinct from the general phenotype or genotype of individuals in a population.
The main reason behind genetic drift is that a subsample, i.e., an isolated or small population, is derived from a large sample or population that is not a natural representative of a larger dataset. Mostly, genetic drift occurs in small populations, where alleles that occur infrequently are at a higher risk of being lost. This drift is non-directional and causes various events, including elevated homozygosity of individuals and loss of genetic variation from the population. Large populations are vulnerable to genetic drift via bottleneck or founder effects.
Once the process of genetic drift starts, it will continue till the involved allele is lost by a population or it is the only allele present in a population at a particular locus. This causes a decline in genetic diversity in a population. Compared to a small population, a significant effect of genetic drift in a large, unthreatened population takes longer to be observed.
Genetic drift plays a crucial role in conservation biology, associated with the determination of the minimal viable population size of a species. It also plays a vital role in the evolution of new species because it can result in a new population being genetically distinct from its original population.
Another consequence of genetic drift is inbreeding, which occurs because of the mating between two genetically similar organisms. Inbreeding increases the risk of an individual inheriting two harmful recessive alleles from both parents, which were low in frequency in the population before drift occurred.
Types of Genetic Drift
The two common types of genetic drift are bottlenecks and the founder effect. These are discussed below:
Bottleneck Effect
Genetic drift could happen due to natural events or disasters that lead to a large number of deaths in a population, randomly. The bottleneck effect manifests when only a few individuals survive in a population. This leads to a reduction in variation in the gene pool of a population. The genetic structure of surviving individuals ultimately becomes the genetic structure of the entire population, which could be significantly different from the original population before the natural disaster.
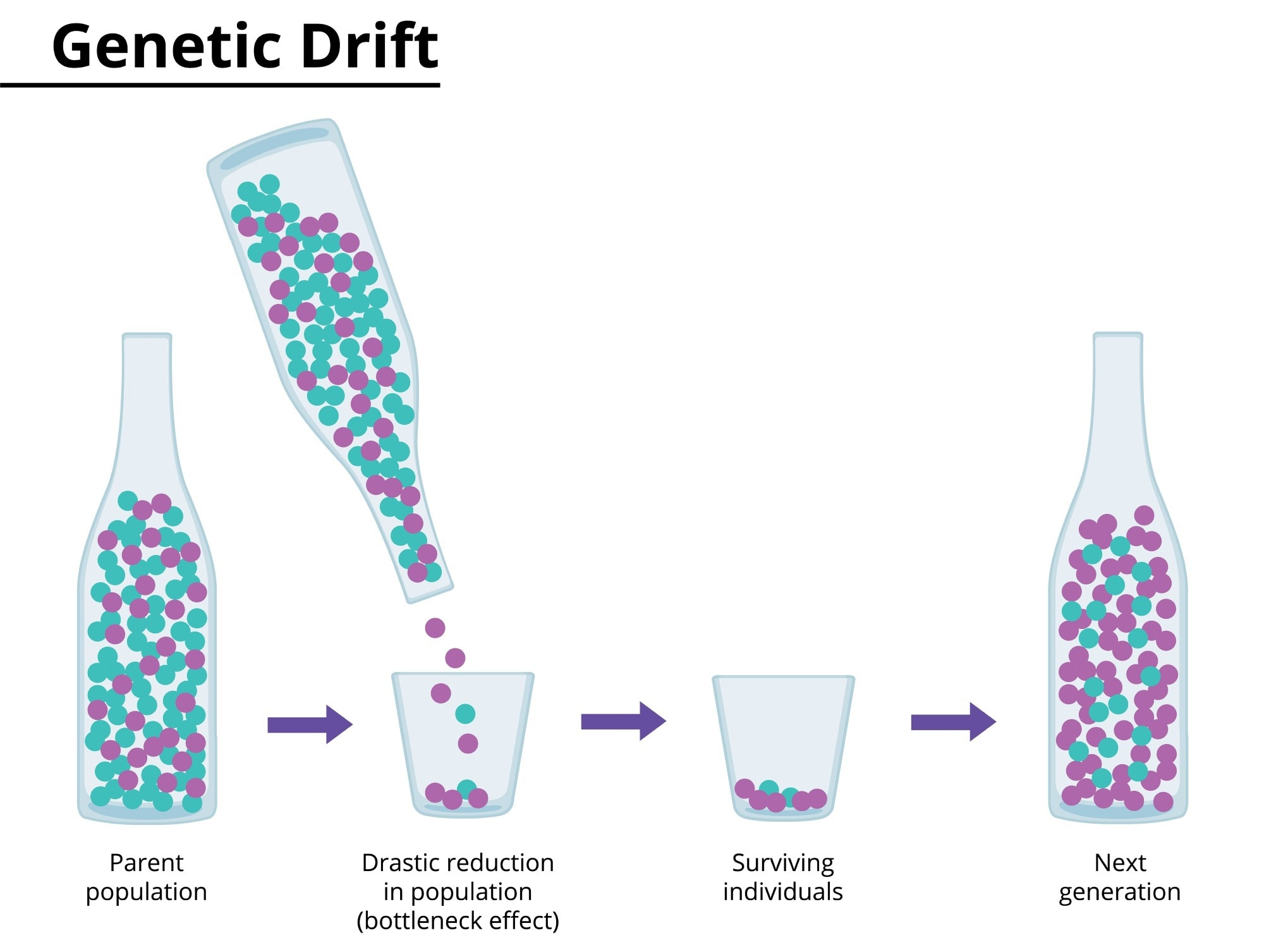
Image Credit: angela_cora/Shutterstock.com
Founder Effect
The founder effect occurs when a population experiences genetic drift because some portion of the population leaves to start a new population in a new geographical region. Also, when a population gets divided due to some physical barrier, the founder effect occurs. In these cases, individuals are not true representatives of the original population. The founder effect occurs when the genetic structure is modified to fit into the new population's founding fathers and mothers.
Estimation of Genetic Drift Based on Effective Population Size
Effective population size (Ne) indicates the sensitivity of a population to genetic drift. Besides population size, other factors that influence the change of allele frequencies over time are an uneven sex ratio, non-random variation in family size, and past fluctuations in population size. Genetic drift can be estimated based on Ne and the exclusion of other aforementioned factors.
Ne can be empirically estimated over short- and long-term time scales. Based on these estimates, it has been observed that small populations are at a higher risk of maladaptation, such as a mismatch between environment and mean phenotype, compared to a large population. Furthermore, the chances of accumulation of mildly deleterious mutations in a small population are significantly higher as selection cannot effectively remove them. This leads to the establishment of deleterious mutations, subsequently influencing population growth rate and size.
Real-world Examples of Genetic Drift
The founder effect plays a vital role in the genetic history of the Afrikaner population of Dutch settlers in South Africa. This finding is based on the presence of specific mutations that are commonly found only in Afrikaners but rarely in other populations. Since a higher-than-normal proportion of the founding individuals carried these mutations, this population expresses a significant incidence of Fanconi anemia (FA) and Huntington's disease (HD).
The founder effect has also been linked to the incidence of Ellis-van Creveld syndrome, a form of dwarfism, in the Amish population of Lancaster County, Pennsylvania, for decades. This syndrome was traced back to Samuel King and his wife, who came to the Amish region in 1744. The mutated genes were passed from the Kings to their offspring. Currently, this syndrome is more commonly found in the Amish population than the American population.
Cheetahs are presently found in a small fraction across eastern and southern Africa and Asia. This species has been classified as endangered by the IUCN Red List. Two genetic drift events had occurred in ancestral populations. The first one was the founder effect that occurred when cheetahs migrated to Africa and Eurasia from the Americas around 100,000 years ago. The second genetic drift event was a bottleneck effect that coincided with large mammal extinctions in the Late Pleistocene.
Sources
- Chiu, C. et al. (2023) Environmental effects, gene flow and genetic drift: Unequal influences on genetic structure across landscapes. Journal of Biogeography. 50(2), pp. 352-364. https://doi.org/10.1111/jbi.14537
- Willi, Y. et al. (2022) Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proceedings of the National Academy of Sciences. 119(1), e2105076119. https://doi.org/10.1073/pnas.2105076119
- Lynch, M. et al. (2016) Genetic drift, selection and the evolution of the mutation rate. Nature Reviews Genetics. 17(11), pp. 704-714. https://doi.org/10.1038/nrg.2016.104
- Hays, C. and Fagan, C. (2016) Conservation Biology, Evolution and. Encyclopedia of Evolutionary Biology. pp. 347-353. https://doi.org/10.1016/B978-0-12-800049-6.00301-2
- Genetic Drift. (2016). [Online] Available at: https://plato.stanford.edu/entries/genetic-drift/
- Honnay, O. (2013). Genetic Drift. Brenner's Encyclopedia of Genetics (Second Edition). pp. 251-253. https://doi.org/10.1016/B978-0-12-374984-0.00616-1
- Britannica, T. Editors of Encyclopaedia (2012). deme. Encyclopedia Britannica. https://www.britannica.com/science/deme-biology
- Masel, J. (2011) Genetic Drift. Current biology. 21(20), pp. R837–R838. https://doi.org/10.1016/j.cub.2011.08.007
Further Reading