For researchers investigating the regulatory processes in cells vital to all life, an innovative system that amplifies gene expression signals could be a turning point.
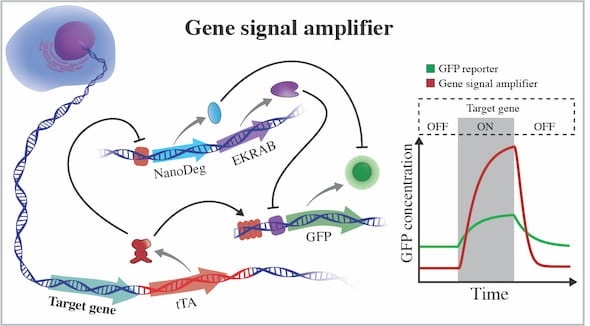
The gene signal amplifier developed by bioscientists at Rice excels at detecting the expression of target genes and can also be used to detect potentially any cellular gene. The amplifier is linked to a cell’s chromosome and directly reports on the activity of a gene by expressing fluorescent proteins (GFP). When the gene is not active, the amplifier expresses negative regulators that quench GFP by operating at different hierarchical levels of cellular information flow. EKRAB is a transcriptional repressor and NanoDeg is a post-translational regulator. When the gene is active, tTA produces GFP and blocks expression of the negative regulators. Image Credit: Segatori Research Group.
A versatile gene signal amplifier has been created at the Rice University laboratory of bioscientist Laura Segatori. When compared to existing methods, the new amplifier can better detect the expression of target genes.
The researchers believe that eventually, the two-module system will enable a simple diagnosis of diseases such as diabetes, Alzheimer’s, and certain types of cancers characterized by unique patterns of protein expression. According to researchers, it could also allow cell-based therapies through which diseased cells could synthesize their own medicine at the point of need.
The study has been reported in Nature Chemical Biology.
The first module forms a part of a chain of synthetic genetic code integrated into a mammalian cell’s DNA by CRISPR-Cas9 editing. Upon adding next to a target gene, the code activates a genetic circuit tracking the gene and, whenever the gene synthesizes a protein, the circuit also discharges a green fluorescent protein (GFP).
The circuit has been designed so that it amplifies the GFP signal, facilitating the tracking of even slight changes in the target gene, which cannot be detected using existing tools.
When the gene turns inactive, the second module developed from an antibody first detected in camels inhibits the synthesis of the fluorescent protein and impairs any nearby GFPs. The combination offers scientists a robust “on-off” signal that also responds to the dynamics of the target gene expression.
The circuit triggers the GFP expression when there is an increase in the gene expression while also preventing the expression of negative regulators of GFP, for example, the nanobody.
Being able to monitor gene expression with high sensitivity is really important for a variety of biomedical applications. It’s important to have a detection system that is sensitive to even small changes in gene expression, which are often biologically relevant. It is also critical to a detection system that provides good dynamic resolution so we can follow gene expression dynamics, which are typically a key determinant of cell behavior.”
Laura Segatori, Associate Professor of Bioengineering, Chemical and Biomolecular Engineering, and Biosciences, Rice University
“That’s what our gene signal amplifier essentially does,” Segatori added. “We developed a genetic circuit that, first of all, we can link to any gene in the chromosome, thus generating a tool that recapitulates the chromosomal context with all the associated complexity of regulation. We don’t have any type of extrachromosomal reporters. This approach provides a sensitive way to monitor all the regulatory and epigenetic mechanisms that regulate gene expression.”
Then we developed a method to amplify the signal so we’re able to monitor really small changes in expression. It’s very robust and stable and has high dynamic resolution.”
Laura Segatori, Associate Professor of Bioengineering, Chemical and Biomolecular Engineering, and Biosciences, Rice University
According to Segatori, it is possible to tweak the system to potentially monitor cellular genes of any type. “We can create multiplex reporter systems for monitoring group of genes that are relevant to the development of a certain disease or that provide a comprehensive readout for a certain signaling pathway or phenotype,” she added.
The technique was demonstrated by the researchers on a range of cells, and a multiplex reporter was produced to monitor markers related to three signaling pathways sensitive to stress in the endoplasmic reticulum of a mammalian cell. The circuit was found to sufficiently improve the fluorescent signal, identifying even slight changes in expression.
Launched by the Rice lab in 2017, the second module is a NanoDeg circuit that serves as a post-translation control offering the system its extensive dynamic range, added Segatori.
Under basal conditions, the circuit expresses not only a transcriptional regulator that inhibits expression of GFP but also NanoDeg molecules that degrade any GFP present in the system, so the cell goes completely dark. And we can tune the system to adapt it to detection of genes with different basal expression by using appropriate doses of inducers of the circuit components.”
Laura Segatori, Associate Professor of Bioengineering, Chemical and Biomolecular Engineering, and Biosciences, Rice University
Through experiments, it was verified that when the system is integrated into the chromosome of the cell, it had no impact on the expression of target genes.
During the study, the lab also created a mathematical model that can be used by researchers to tailor the amplifier platform to track any target gene and estimate the ideal doses of small-molecule inducers employed for gene expression regulation.
Segatori and her colleagues are making efforts to optimize the platform, which was developed by Rice graduate student and lead author Carlos Origel Marmolejo, mainly to treat diseases.
“There is a great interest currently in developing cell therapies that are feedback-responsive,” noted Segatori. “Our platform could allow production of therapeutics in response to detection of gene expression signatures relevant to a certain disease or environmental condition.”
Graduate student Bhagyashree Bachhav and undergraduates Sahiti Patibandla and Alexander Yang are the co-authors of the study. Segatori is an associate professor of bioengineering, chemical and biomolecular engineering and biosciences.
The National Science Foundation and the Welch Foundation supported the research.
Source:
Journal reference:
Origel Marmolejo, C. A., et al. (2020) A gene signal amplifier platform for monitoring the unfolded protein response. Nature Chemical Biology. doi.org/10.1038/s41589-020-0497-x.