Chemists from MIT and Harvard University have elucidated the structure of a unique bacterial enzyme with the ability to break down an amino acid found in collagen.
Collagen is the most abundant protein found in the human body.
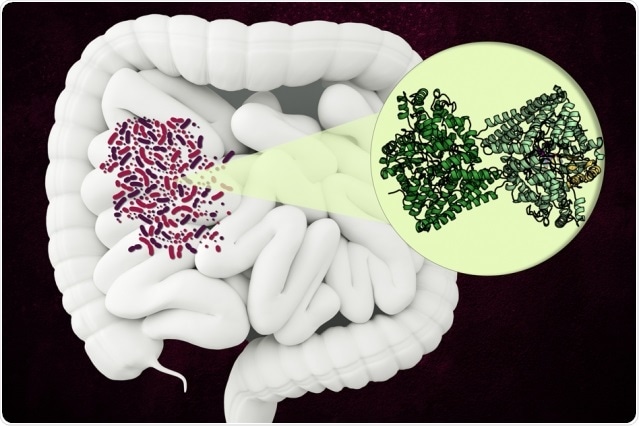
MIT researchers have discovered the structure of an unusual enzyme that some microbes use to break down a component of collagen in the human gut. Image Credit: Christine Daniloff, MIT.
Named hydroxyl-L-proline dehydratase (HypD), the enzyme is found in a few hundred types of bacteria that reside in the human gut, such as Clostridioides difficile.
The enzyme induces a new type of chemical reaction that disintegrates hydroxy-L-proline—the molecule responsible for the hard and triple-helix structure of collagen.
By knowing the structure of the enzyme, the researchers can develop drugs that block it, which would be helpful in the treatment of C. difficile infections that have become resistant to several existing antibiotics.
This is very exciting because this enzyme doesn’t exist in humans, so it could be a potential target. If you could potentially inhibit that enzyme, that could be a unique antibiotic.”
Catherine Drennan, Professor of Chemistry and Biology, MIT
Drennan is also an investigator at Howard Hughes Medical Institute. The senior authors of the study, which was recently reported in the eLife journal, are Drennan and Emily Balskus, a professor of chemistry and chemical biology at Harvard University.
The lead authors of the study are MIT graduate student Lindsey Backman and former Harvard graduate student Yolanda Huang.
A difficult reaction
The HypD enzyme belongs to a large class of proteins known as glycyl radical enzymes that work unusually by transforming a molecule of glycine—the simplest amino acid—into a radical, a molecule with an unpaired electron.
Since these radicals are highly unstable and reactive, they are used as cofactors that help initiate challenging chemical reactions.
These enzymes function best in conditions deprived of oxygen, like the human gut. As part of the Human Microbiome Project, which has sequenced thousands of bacterial genes from various species located in the human gut, it has produced various types of glycyl radical enzymes, including HypD.
In earlier research work, Balskus and colleagues from the Broad Institute of MIT and Harvard identified that HypD can disintegrate hydroxyl-L-proline into a precursor of proline—one of the essential amino acids—by eliminating the hydroxyl modification as a water molecule.
These bacteria can eventually use proline for ATP generation—a molecule used by cells for storage of energy, by a process named amino acid fermentation.
HypD has been observed in around 360 species of bacteria found in the human gut. In this study, Drennan and her team analyzed the structure of the version of HypD found in C. difficile using X-ray crystallography.
In 2011, these species of bacteria caused nearly half a million infections and 29,000 deaths in the United States.
The team could identify the region of protein which forms the enzyme’s “active site”—the site where the reaction occurs. When hydroxyl-L-proline attaches to the active site, a glycyl radical is formed by an adjacent glycine molecule. This radical can be passed onto the hydroxy-L-proline, causing the elimination of the hydroxyl group.
In general, the removal of the hydroxyl group is a challenging reaction as it requires higher inputs of energy.
By transferring a radical to hydroxy-L-proline, it lowers the energetic barrier and allows for that reaction to occur pretty rapidly. There’s no other known enzyme that can perform this kind of chemistry.”
Lindsey Backman, Graduate Student, MIT
New drug target
When the bacteria carry out this reaction, they seem to redirect proline into their metabolic pathways to help them grow. Thus, by blocking this enzyme, the growth of bacteria could be slowed down.
It could be beneficial in controlling C. difficile, which usually occurs in small numbers in the human gut but could lead to illness if the population becomes very large. At times, it occurs after antibiotic treatment that eliminates other species and enables the proliferation of C. difficile.
C. difficile can be in your gut without causing problems—it’s when you have too much of it compared to other bacteria that it becomes more problematic. So, the idea is that by targeting this enzyme, you could limit the resources of C. difficile, without necessarily killing it.”
Catherine Drennan, Professor of Chemistry and Biology, MIT
The scientists expect to start the designing of drug candidates with the potential to block HypD, by targeting the protein structure elements that play a vital role in performing its function.
The study was financially supported by the National Institutes of Health, National Science Foundation Graduate Research Fellowship, Harvard University, a Packard Fellowship for Science and Engineering, the NSERC Postgraduate Scholarship-Doctoral Program, an Arnold O. Beckman Postdoctoral Fellowship, a Dow Fellowship, and a Gilliam Fellowship from the Howard Hughes Medical Institute.
Source:
Journal reference:
Backman, L. R. F., et al. (2020) Molecular basis for catabolism of the abundant metabolite trans-4-hydroxy-L-proline by a microbial glycyl radical enzyme. eLife. doi.org/10.7554/eLife.51420.