Scientists in the United States have used a newly developed fluorescent imaging technique to create high-resolution maps of microbial communities on the human tongue.
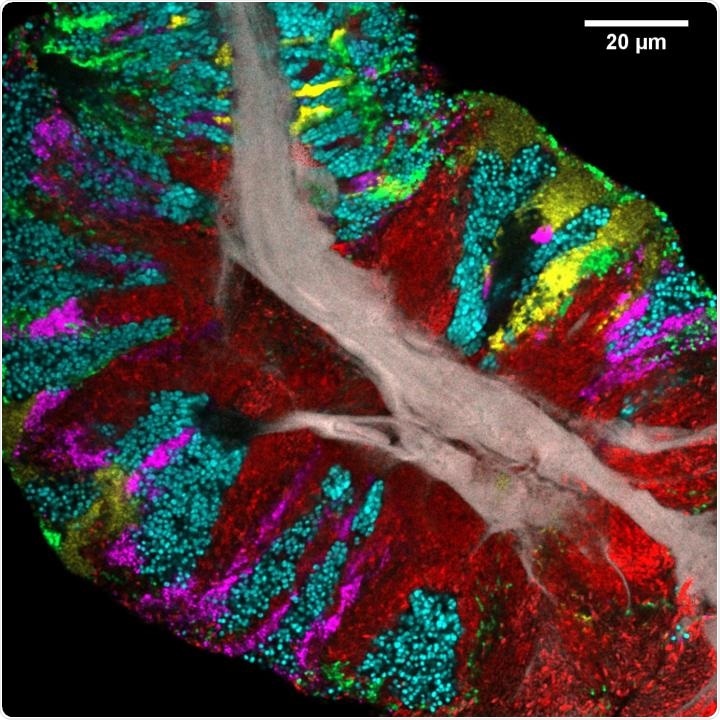
Bacterial biofilm scraped from the surface of the tongue and imaged using CLASI-FISH. Human epithelial tissue forms a central core (gray). Colors indicate different bacteria: Actinomyces (red) occupy a region close to the core; Streptococcus (green) is localized in an exterior crust and in stripes in the interior. Other taxa (Rothia, cyan; Neisseria, yellow; Veillonella, magenta) are present in clusters and stripes that suggest the growth of the community outward from the central core. Image credit: Steven Wilbert and Gary Borisy, The Forsyth Institute.
Bacterial biofilm scraped from the surface of the tongue and imaged using CLASI-FISH. Human epithelial tissue forms a central core (gray). Colors indicate different bacteria: Actinomyces (red) occupy a region close to the core; Streptococcus (green) is localized in an exterior crust and in stripes in the interior. Other taxa (Rothia, cyan; Neisseria, yellow; Veillonella, magenta) are present in clusters and stripes that suggest the growth of the community outward from the central core. Image credit: Steven Wilbert and Gary Borisy, The Forsyth Institute.
The images, which were published in the Cell Reports journal on 24th March, show that the microbial biofilms found on the surface of the tongue have an intricate, highly ordered spatial arrangement.
From detailed analysis of the structure, we can make inferences about the principles of community growth and organization. Bacteria on the tongue are a lot more than just a random pile. They are more like an organ of our bodies.”
Gary Borisy, Study Senior Author, Forsyth Institute and the Harvard School of Dental Medicine
The human oral microbiome is an intricate ecosystem. A range of factors influence the spatial arrangement of microbial communities in the mouth, such as frequency of disturbances (like an abrasion or oral hygiene), moisture, temperature, oxygen, pH, and salivary flow.
Moreover, microbes have an impact on their neighbors by serving as sources and sinks of nutrients, metabolites, and suppressive molecules like antimicrobial peptides and hydrogen peroxide.
Microbes can occupy space and physically exclude each other from preferable habitats, but their surfaces include binding sites to which other microbes could attach themselves. However, in the field of microbial ecology, spatial patterning has not gained considerable attention.
We think that learning who is next to who will help us understand how these communities work. The tongue is particularly important because it harbors a large reservoir of microbes and is a traditional reference point in medicine. ‘Stick out your tongue’ is one of the first things a doctor says.”
Jessica Mark Welch, Study Co-Author and Microbial Ecologist, Marine Biological Laboratory, Woods Hole, Massachusetts
In this study, the team employed a technique named Combinatorial Labeling and Spectral Imaging–Fluorescence in situ Hybridization (CLASI-FISH), which was newly developed in the Borisy lab.
In this approach, a specified type of microorganism is labeled with multiple fluorophores, which increases the types of microbes that can be simultaneously identified and localized in a single field of view.
Our study is novel because no one before has been able to look at the biofilm on the tongue in a way that distinguishes all the different bacteria, so that we can see how they arrange themselves.”
Gary Borisy, Study Senior Author, Forsyth Institute and the Harvard School of Dental Medicine
Borisy added, “Most of the previous work on bacterial communities used DNA sequencing-based approaches, but to get the DNA sequence, you have to first grind up the sample and extract the DNA, which destroys all the beautiful spatial structure that was there. Imaging with our CLASI-FISH technique lets us preserve the spatial structure and identify the bacteria at the same time.”
Firstly, analyzed sequence data was used to detect major bacterial taxa present in small samples scraped from the tongues of 21 healthy subjects.
With guidance from sequence analysis, the imaging method targeted major genera and specific species to achieve an overall view of the microbiome structure. The team identified 17 bacterial genera that largely found on the tongue and present in over 80% of the subjects.
The samples included bacteria attached to host epithelial cells, free bacteria, and bacteria organized into consortia, which are multi-layer biofilms with a complex structure.
The consortia revealed inconsistency in community structure, including spatially localized domains in which a single taxon was highly predominant. Although they differed in shape, their length was usually tens to hundreds of microns, with a core of epithelial cells and a clearly defined perimeter.
The tongues of all individuals contained consortia comprising three genera, which are Actinomyces, Rothia, and Streptococcus. Actinomyces was often observed near the core, whereas Rothia usually appeared as large patches toward the exterior of the consortium. It was found that Streptococcus form veins or patches in the interior of the consortia and a thin crust on the exterior.
According to Mark Welch, “ Collectively, our species-level imaging results confirm and deepen our understanding of habitat specificity of key players and show the value of investigating microbiomes at high imaging and identification resolution.”
The study outcomes collectively propose a model for how the ordered microbial communities found on the tongues of the subjects were generated. Initially, the bacterial cells bind to the surface of the tongue’s epithelium either individually or as small clusters.
Due to the increase in population, varied taxa push on one another and multiply quickly in microenvironments that aid their physiological requirements. This differential proliferation leads to the patch mosaic arrangement seen in larger and more mature structures.
The images also uncovered the fact that certain taxa capable of nitrate reduction—such as Neisseria, Actinomyces, Veillonella, and Rothia—are predominantly found in tongue consortia.
This increases the possibility that small projections that erupt on the surface of the human tongue are structured to support the growth of bacteria that transform salivary nitrate to nitrite—a function that is not encoded by the human host genome.
Source:
Journal reference:
Wilbert, S. A., et al. (2020) Spatial Ecology of the Human Tongue Dorsum Microbiome. Cell Reports. doi.org/10.1016/j.celrep.2020.02.097.