Scientists at the University of Illinois at Urbana-Champaign have created the first-ever computational model of a human cell and simulated its behavior for 15 minutes.
This is the longest time realized for such a complex biological system.
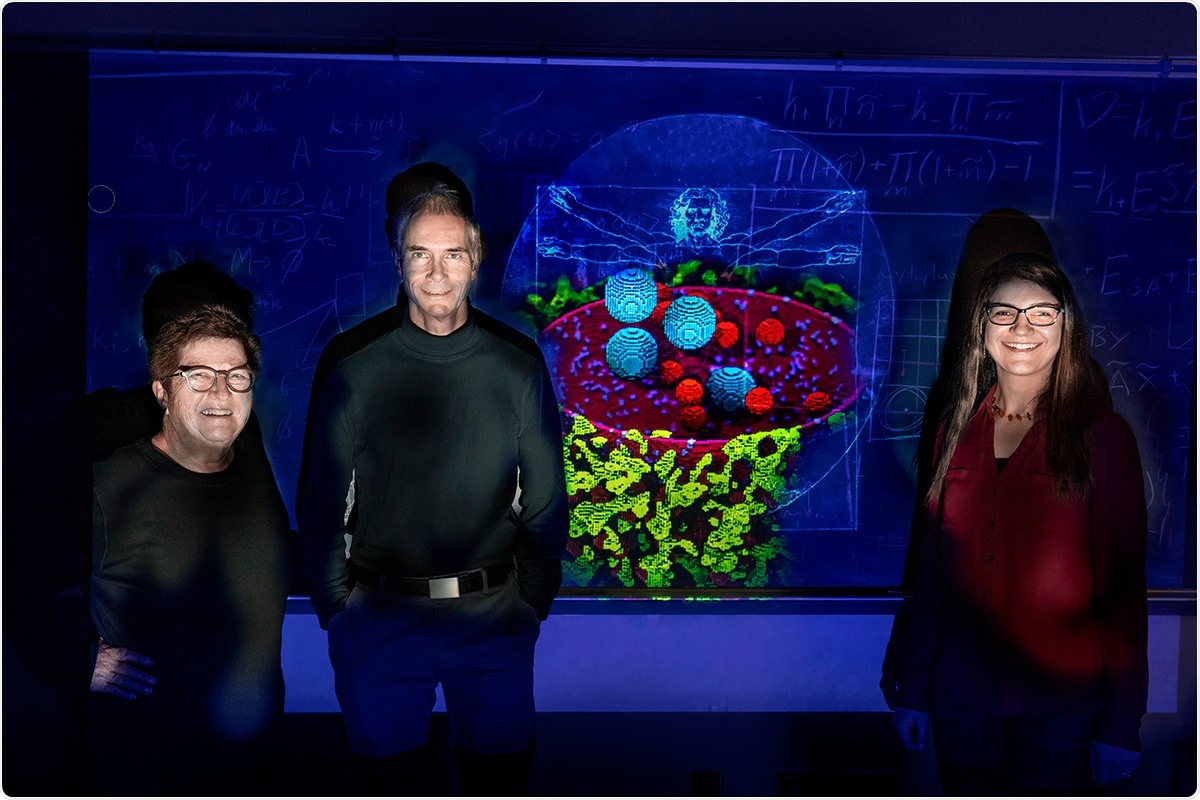
Chemistry professors Zaida Luthey-Schulten, left, Martin Gruebele, and research scientist Zhaleh Ghaemi have developed the most complete computational model of a human cell to date. Image Credit: Fred Zwicky.
Simulations performed as part of their new study demonstrate the impacts of spatial arrangement inside the cells on certain genetic processes manipulating the regulation and development of human traits and specific human diseases.
The study reported in the PLoS Computational Biology journal presents a new computational platform that can be accessed by all researchers.
This is the first program that allows researchers to set up a virtual human cell and change chemical reactions and geometries to observe cellular processes in real time.”
Zhaleh Ghaemi, Study Lead Author and Research Scientist, University of Illinois at Urbana-Champaign
Based on the concept that cells’ interior is packed with different molecules and organelles, Zaida Luthey-Schulten, a chemistry professor, and her team focused on how the movement of individual molecules around different obstacles impact the chemical reactions within cells.
The researchers tested the new model by performing simulations of a process known as RNA splicing—one of the most complex cellular processes characteristic to human cellular biology.
RNA splicing changes the messenger RNA molecules that carry information needed from DNA to form proteins. The process uses a complex cellular machine—called a spliceosome—that requires the trafficking of precursor and mature components around the highly compartmentalized parts of a cell. This makes RNA splicing ideal for studying how spatial arrangement affects the various chemical reactions that take place in cells.”
Zhaleh Ghaemi, Study Lead Author and Research Scientist, University of Illinois at Urbana-Champaign
According to the researchers, the new simulations demonstrated why precursors of the spliceosome tend to move between the nucleus and compartments of cytoplasm.
Even though this movement seems somewhat inefficient and counterintuitive at first glance, our simulations indicate that they are essential to the proper RNA splicing, and therefore protein synthesis. When protein synthesis goes awry, it can lead to disease, including cancer.”
Martin Gruebele, Study Co-Author and Chemistry Professor, University of Illinois
The computational platform was designed to model a range of cellular processes and to be fully customizable by the researcher who uses it.
According to Luthey-Schulten, “For example, we could use this model to observe what types of proteins will form if the RNA-splicing process were to remove only two parts of a DNA sequence instead of three. This could provide insights into how different proteins form and influence the development of cancer cells.”
The researchers said that even though their computational model was one of the most detailed human cell models developed until now, there is more scope for advancing and tailoring it to study other cellular processes.
Gruebele added, “This simulation allowed us to observe the RNA splicing for 15 minutes. Ultimately, we would like to be able to run the program for much longer and include all of the proteins that are required for gene replication, allowing us to observe cell division in real-time. The possibilities for our group—and others because the program is open access—are endless.”
Source:
Journal reference:
Ghaemi, Z., et al. (2020) An in-silico human cell model reveals the influence of spatial organization on RNA splicing. PLoS Computational Biology. doi.org/10.1371/journal.pcbi.1007717.