The Structural Biology Lab at Kazan Federal University and the Institute of Proteins at the Russian Academy of Sciences represent the Russian side for the current study.
This specific research deals with the problem of stress resistance found in Staphylococcus aureus. The outcomes can help in identifying novel antibiotics.
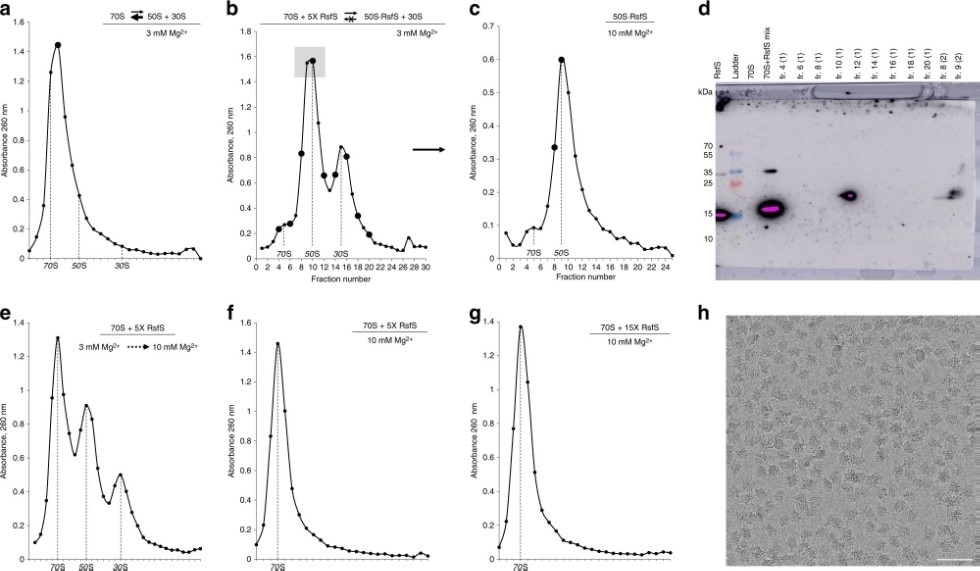
RsfS protein binds to the 50S particles and prevents ribosomal subunits association. Image Credit: Kazan Federal University.
Konstantin Usachev, head of Structural Biology Lab, explained that the study was the outcome of a five-year-long collaboration between Kazan Federal University, Stuttgart University, Institute of Proteins, and the Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France.
Back in 2016, the researchers were the first to fully elucidate the structure of the ribosome in Staphylococcus aureus and compare it to other kinds of organisms.
The ribosome is the largest ribonucleic complex in the cell, and it consists of two subunits: large and small. The small subunit is responsible for reading the genetic code, and the function of the large subunit is to ensure the formation of the peptide bond in the growing protein chain.”
Konstantin Usachev, Head of Structural Biology Lab, Kazan Federal University
Usachev continued, “In our article, using cryoelectron microscopy and X-ray diffraction methods, we were able to show the ribosome binding mechanism of the RsfS protein (Ribosome silencing factor S), which protects Staphylococcus aureus from stress (antibiotics, fever, or host immunity). Under stress, this protein binds to the large subunit of the ribosome and prevents the small subunit from joining, preventing the formation of functional ribosomes.”
Studies on this topic were started five years ago. For a long period, researchers were unable to acquire the structure of the intricate RsfS protein-bacterial ribosome complexes in excellent resolution, which was essential to interpreting the specifics of their mechanism of action.
One of the problems was the high toxicity of this protein to the cells of the bacteria E. coli, the organism used to produce proteins in the laboratory. The fact is that the RsfS protein, which stops the synthesis of proteins in Staphylococcus aureus, is able to stop this process in other bacteria. This resulted in a very small amount of protein sample, insufficient for structural studies.”
Konstantin Usachev, Head of Structural Biology Lab, Kazan Federal University
Usachev continued, “In addition, the sample was extremely unstable and aggregated. Then the idea came to us—to isolate this protein simultaneously with its target in the structure of staphylococcus ribosome—protein L14, which is a part of the large subunit. It turned out that if you select both components at the same time, they will be stable in solution.”
“We were able to obtain crystals of these proteins and solve the structure by X-ray diffraction analysis, first with medium resolution using the new single-crystal diffractometer available in our laboratory, and then with high resolution using the ESRF synchrotron in Grenoble, France,” Usachev further added.
The researchers have to subsequently analyze the details of the interaction between the RsfS protein and the ribosome of Staphylococcus aureus. This feat could be realized through cryoelectron microscopy.
Unfortunately, we did not have a microscope capable of solving high-resolution structures using this method, but NovAliX became interested in our studies and offered their microscope to test the initial samples. The obtained samples of the complexes had turned out so good that the company then contacted one of the world’s leading manufacturers of FEI microscopes in the Netherlands and organized data collection on a Titan Krios microscope.”
Konstantin Usachev, Head of Structural Biology Lab, Kazan Federal University
“As a result, combining the data of cryoelectron microscopy with the previously obtained data of X-ray diffraction analysis, we were able to show in detail the molecular mechanism of the action of the RsfS protein on Staphylococcus aureus ribosomes,” Usachev concluded.
The Structural Biology Lab has now teamed up with the Chemoinformatics Lab to identify the structure of promising antibiotics, which can disrupt the functioning of RsfS protein, and thus successfully eliminate Staphylococcus aureus.
Usachev informed that only the collective efforts of physicists, chemists, and biologists could improve the studies in protein synthesis and the development of novel medications.
Source:
Journal reference:
Khusainov, I., et al. (2020) Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach. Nature Communications. doi.org/10.1038/s41467-020-15517-0.