Autophagy can be described as a housekeeping process using which cells eliminate dysfunctional contents to stabilize the energy sources during stressful situations.
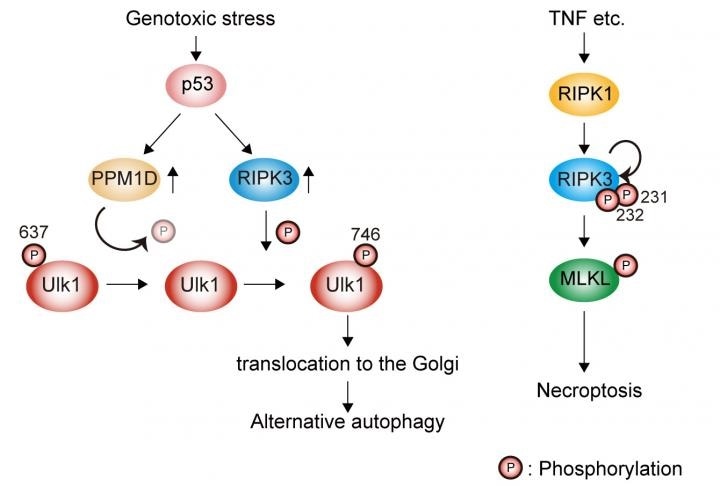
Genotoxic stress induces Ulk1 dephosphorylation at Ser637 in a p53/PPM1D-dependent manner. The dephosphorylated Ulk1 is then phosphorylated at Ser746 by a necroptosis initiator, RIPK3, and translocates to the Golgi, which results in alternative autophagy (left). TNF signaling induces necroptosis but not alternative autophagy (right). Image Credit: Department of Pathological Cell Biology, Medical Research Institute, Tokyo Medical and Dental University.
Scientists from Tokyo Medical and Dental University (TMDU) have now discovered a unique molecular mechanism through which a type of autophagy, known as alternative autophagy, is stimulated.
In the latest research published in the Nature Communications journal, the researchers demonstrated how a particular phosphorylation site of the protein Unc51-like kinase 1 (Ulk1) is crucial for the cells to travel the alternative autophagy route.
Cells are living structures and guarantee homeostasis by performing certain processes through which they construct and degrade their contents. Specifically, during stressful situations, for instance, at the time of exposure to toxins, autophagy helps to guarantee an organized turnover procedure through which cells can survive by recycling their contents.
Fascinatingly, the autophagy process can occur through many distinct molecular mechanisms, and alternative autophagy and canonical autophagy are two such mechanisms.
While the Ulk1 protein is known to trigger both kinds of autophagy, the mechanism through which this protein differentially controls them has continued to be elusive.
Autophagy is a very elaborate process by which cells recycle their contents. The goal of our study was to understand how Ulk1 that has control over two types of autophagy, differentially regulates them.”
Shigeomi Shimizu, Study Corresponding Author, Tokyo Medical and Dental University
To realize their objective, the scientists employed mouse embryonic fibroblasts (MEFs) that lacked the Atg5 protein to switch off the canonical autophagy. The team subsequently induced alternative autophagy by subjecting these MEFs to a DNA-damaging reagent called etoposide.
With the help of mass spectrometry, the scientists discovered that the Ulk1 protein carried an extra phosphoryl group at its amino acid serine in position 746 (Ser746; p-Ulk1746), also referred to as phosphorylation, when subjected to the etoposide treatment but not when left untreated.
By creating a novel antibody against p-Ulk1746, the scientists subsequently demonstrated that the protein is localized to the Golgi complex inside the cells. The Golgi complex is a kind of organelle that is involved in several cellular processes, such as the alternative autophagy.
While these were already exciting findings, our goal was to understand whether the specific phosphorylation of Ulk1 at the serine 746 site is required for alternative autophagy and which kinase is responsible for this phosphorylation step.”
Satoru Torii, Study Corresponding Author and Study Lead, Tokyo Medical and Dental University
To study the causal association between alternative autophagy and Ulk1 Ser746 phosphorylation, the scientists employed a fluorescent tandem protein-containing green fluorescent protein (GFP) and red fluorescent protein (RFP).
GFP cannot fluoresce within acidic settings, and therefore, the tandem protein makes the autolysosomes turn red. Autolysosomes are cellular compartments that are produced at the time of autophagy.
Although the red fluorescence emerged after the etoposide treatment, it was not produced in cells that generate the Ulk1 nonphosphorylated mutant. This implies that p-Ulk1746 is needed for alternative autophagy.
The scientists then demonstrated that a protein called receptor-interacting protein kinase 3 (RIPK3), which phosphorylates other types of proteins implicated in necroptosis, accounts for the production of p-Ulk1746 by demonstrating that alternative autophagy and p-Ulk1746 actually took place in normal cells but not in RIPK3-deficient cells.
Interestingly, in Atg5-expressing MEFs, RIPK3-deficiency did not impact canonical autophagy, implying that p-Ulk1746 does not play a role in canonical autophagy.
These are striking results that shed new light on how cells regulate the complex process of autophagy. We hope that our findings will be helpful in understanding the role of alternative autophagy in normal biology and disease.”
Shigeomi Shimizu, Study Corresponding Author, Tokyo Medical and Dental University
Source:
Journal reference:
Torii, S., et al. (2020) Identification of a phosphorylation site on Ulk1 required for genotoxic stress-induced alternative autophagy. Nature Communications. doi.org/10.1038/s41467-020-15577-2.