Scientists from the Quadram Institute and Earlham Institute have developed a unique tool that is helping them to translate the complex communication that takes place between the body and the microbiome.
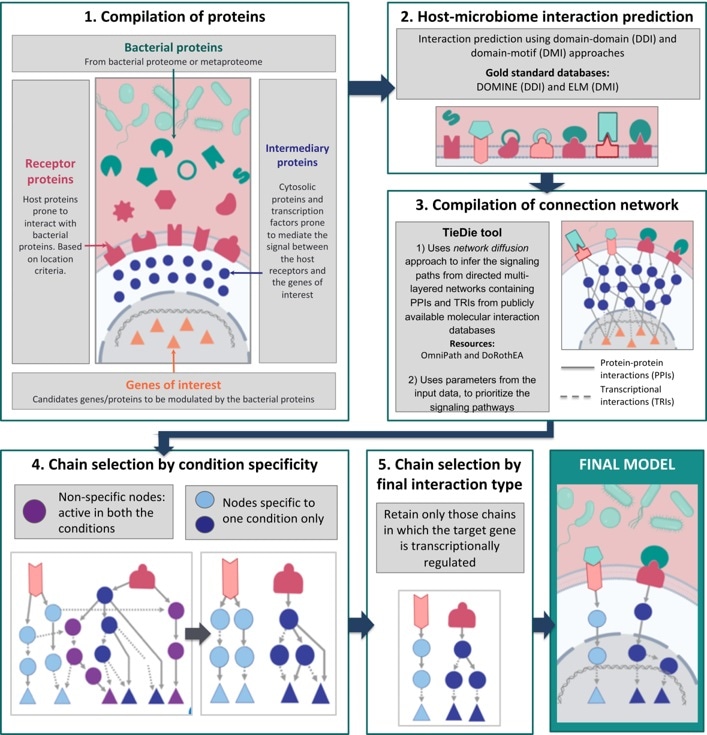
The MicrobioLink workflow. Image Credit: Quadram Institute.
The tool could be useful for scientists who are trying to figure out the effect of microbes on health, the changes that cause the disease, and point to promising drug targets.
The human gut microbiome plays significant roles in keeping the individuals healthy. It is important for absorbing nutrients from food, and it is becoming more evident that it plays a major role in different terminal, chronic, and lifestyle diseases.
Variations in the composition of the microbiome have been linked to inflammatory bowel disease (IBD), obesity, and type 2 diabetes, but it is uncertain how those variations cause the onset of various conditions, or whether they are actually a symptom.
Scientists have been targeting the communication between the microorganisms and their host organisms, including humans, to expose the mechanisms. Microbes discharge a wide range of RNA molecules, proteins, and metabolites that enable them to interact with other proteins from the host organism. This may trigger a response from the host networks, leading to switching on or off the host genes that modulate numerous processes.
To understand the processes that impact health and disease, it is essential to translate these cross-kingdom interactions between the microbial and host proteins. Such inter-kingdom interaction studies have been performed on a few bacteria, especially pathogenic species that activate specific disease conditions.
However, the microbiome may contain scores of different species, all possibly interacting with a range of different host proteins, associated with several different physiological processes and signaling pathways.
Dr Tamas Korcsmaros and Dr Padhmanand Sudhakar from the Quadram Institute and the Earlham Institute and their collaborators created an integrated computational pipeline known as MicrobioLink to manage these complex communications.
Published in the Cells journal, MicrobioLink links microbial proteins with host proteins they are likely to communicate with, and then deduces how these communications affect the cellular processes in the host.
Scientists can employ Microbiolink to test a group of proteins, which could be the proteins secreted by a whole microbial community, against a pertinent list of host proteins obtained from existing databases or experimental data.
Based on their respective molecular features, like motifs and domains, the pipeline subsequently predicts the communications that take place between the host and microbial proteins. Interactions between the proteins represent potentially fascinating communication channels between the host and bacteria.
The power of Microbiolink appears in the following step that employs the diffusion principle in network science to track the impacts of the communications between the host and microbial proteins on other host processes further downstream.
This involves the use of prevalent databases of validated molecular interactions to track the signals from host proteins (projected to be bound by and thus altered by microbial proteins) by other target proteins or genes in the host.
With suitable filtering, this can detect key nodes in networks and specific proteins or genes in the host affected by the original communication between the host and the microbes.
The researchers performed a case study to test the pipeline. They used a compendium of bacterial proteins that are found only in patients suffering from Crohn’s disease, and another group of proteins present only in healthy individuals.
Using the MicrobioLink, the researchers compared how the different groups of proteins impacted autophagy—a significant cellular process that is known to be dysregulated in Crohn’s disease. This exposed a network with well-defined separated signaling paths, exclusive to the healthy and disease contexts and, eventually, influencing the expression of autophagy genes.
These outcomes would have to be validated experimentally, yet they point toward a possible mechanism connecting the Crohn’s disease and microbiome.
The case study also shows the potential for this integrated computational pipeline to translate the interactions between the host and the microbes so that their effects on the body and their implications on health and disease could be understood.
In addition to studying the effect of the microbiome on other diseases, the MicrobioLink could also be used to unravel the beneficial effects on communities of bacteria or the host of probiotic bacteria, and not just in humans.
Basically, we are now providing a method for the scientific community to understand better how commensal or probiotic bacteria are influencing our health. It is a computational analysis tool, so the first step towards designing focused experimental studies but we hope it will already help colleagues to leverage the existing wealth of data from various patients, and result in new clinical intervention studies.”
Dr Tamas Korcsmaros, Researcher, Quadram Institute
“As MicrobioLink is versatile and agnostic to the organisms, it will have applications in livestock health, as well as in analyzing how the soil microbiome affects plant health,” stated Dr. Padhmanand Sudhakar, a researcher at the Earlham Institute.
In the near future, the researchers are looking to use the MicrobioLink to infer the effects of the SARS-CoV-2 virus behind the COVID-19 infection.
Source:
Journal reference:
Andrighetti, T., et al. (2020) MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions. Cells. doi.org/10.3390/cells9051278.