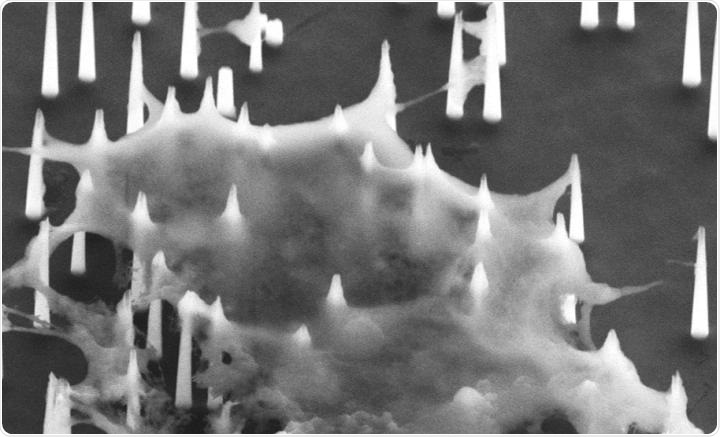
A scanning electron microscope (SEM) image shows the diamond nanoneedles penetrate the cell membrane. Image Credit: Photo source: DOI number: 10.1126/sciadv.aba4971.
To obtain rapid and highly sensitive profiling of miRNAs, a team of researchers from City University of Hong Kong (CityU) has devised a new intracellular biopsy approach that separates the targeted miRNAs from the living cells in less than 10 minutes by using an array of diamond nanoneedles.
This is a simple technique and can even be applied to other aspects, from early cancer diagnosis to the nucleic acid testing of viruses (for example, COVID-19).
The study was headed by Dr. Shi Peng, an Associate Professor of the Department of Biomedical Engineering (BME) at the University. The researchers’ results were recently published in Science Advances, a scientific journal, in a paper titled “High-throughput intracellular biopsy of microRNAs for dissecting the temporal dynamics of cellular heterogeneity.”
Quasi-single-cell miRNA analysis
miRNAs are essentially short non-coding RNA fragments that play a role in the post-transcriptional regulation of gene expression. They strongly control different biological processes, like cell differentiation, survival, and proliferation.
At present, the polymerase chain reaction (PCR) is the most common technique used in miRNA analysis. But this technique is expensive and involves complex processes and also long turnover time owing to procedures like isolation, purification, and amplification of RNAs from cellular lysates.
Moreover, conventional PCR provides only an average measurement for an unlimited number of cells, irrespective of the fact that cells could vary from one another. This problem is particularly significant for detecting rare cells, like cancer stem cells, which are one of the major factors responsible for relapse and metastasis of cancer.
As a substitute, the CityU researchers have developed a high-throughput technique referred to as “inCell-Biopsy,” which is used to considerably streamline the experimental operation in a way that isolation, purification, and amplification of RNAs are no longer needed. The processing time is thus decreased from hours to just about 10 minutes.
The method is based on a “molecular fishing” system that was developed by the team before. In this method, a series of diamond nanoneedles are used as “fishing rods” and P19—a kind of RNA binding protein used as “fishing hooks.”
The P19 RNA can adhere to all the available double-strand RNAs with a length of 19–24 nucleotides. The “fishing rods” were used to perforate the cell membrane and then directly extract the multiple miRNA targets captured by the “fishing hooks” without damaging the cells.
Subsequent observation of the “fishing results” on every nanoneedle thus allows a quasi-single-cell miRNA analysis.
Currently, we can simultaneously pull out 9 miRNA targets in each puncture; the number of targets can be further expanded with more optimization. One nanoneedle patch can be cleaned and reused for more than 50 times.”
Dr Shi Peng, Associate Professor, Department of Biomedical Engineering, City University of Hong Kong
The nanoneedles, which were patented by CityU, were fabricated by Zhang Wenjun, Chair Professor of Materials Science.
Capturing the change of miRNA expression in the same cell population at different time
Unlike PCR-based techniques that involve lysing of cell samples, the cells remain alive even after the nanoscale puncture. Hence, scientists can examine the same types of cells several times to track the changes in the expression of miRNAs related to cellular activity.
This implies that scientists can visualize the evolution of cells by comparing miRNA profiling results. As proof of concept, the scientists applied the inCell-Biopsy method on mouse embryonic stem cells across their differentiation towards motor neurons.
From the results, we can obtain information about the percentage of emerging cell populations originated from the stem cells. We can also tell the relationship between different cell populations. Such analysis was not possible by using existing methods.”
Dr Shi Peng, Associate Professor, Department of Biomedical Engineering, City University of Hong Kong
A potential platform for quality control of therapeutic strategies and beyond
According to Dr. Shi, the intracellular biopsy method would provide scope to quantitatively analyze the temporal dynamics of miRNA expression for the same group of cells to expose the evolution of cellular heterogeneity in mixed populations of cells.
The technique is relevant as a simple and rapid evaluation platform for the quality control of evolving therapeutic methods involving cell components, like the CAR T-cell or stem cell therapies—genetically exploiting a patient’s immune cells to detect and destroy cancer cells. This could improve the safety and effectiveness of the therapies.
Furthermore, miRNA profiling has currently been promoted for earlier detection of cancer in the healthcare sector. The researchers believe that this novel approach could be highly compatible and translational with the present healthcare usage, which renders such an assay considerably shorter and less complicated. The researchers have begun to examine the method in a colon cancer screening study.
Our technique can be used on cells other than stem cells, for example, blood cells, cancer cells or T-cells, etc. And it can be used not only for profiling miRNA, but also for different kinds of RNAs, or even applicable to the detection of COVID-19 viruses.”
Dr Shi Peng, Associate Professor, Department of Biomedical Engineering, City University of Hong Kong
Source:
Journal reference:
Wang, Z., et al. (2020) High-throughput intracellular biopsy of microRNAs for dissecting the temporal dynamics of cellular heterogeneity. Science Advances. doi.org/10.1126/sciadv.aba4971