A team of researchers, headed by Professor XU Guowang from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences, has come up with a procedure through which the pseudotargeted metabolomics technique can be systematically summarized and upgraded.
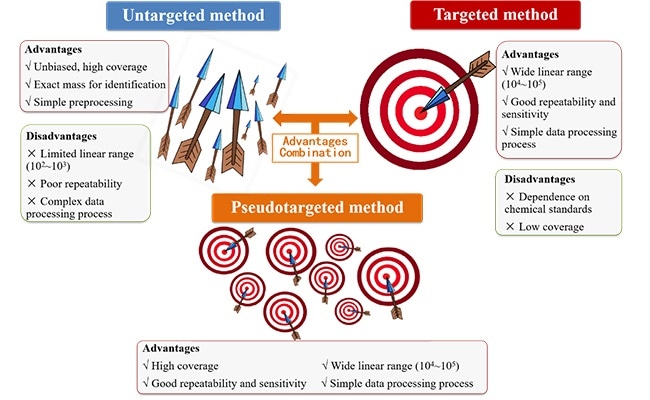
Pseudotargeted metabolomics method is a new method integrating the advantages of untargeted and targeted metabolomics methods. Image Credit: WANG Ting.
This latest study was published in the Nature Protocols journal on June 24th, 2020.
Metabolomics can be described as the science of exploring endogenous metabolites. One of the most predominantly used analytical tools is ultra-high performance liquid chromatography-mass spectrometry. As such, there are two major kinds of analysis strategies—targeted method and untargeted method.
The targeted method employs triple quadrupole mass spectrometry (TQMS). While the data quality is enhanced, only familiar metabolites can be identified. Similarly, the untargeted approach makes use of high-resolution mass spectrometry (HRMS) to acquire extensive metabolite data, with the drawbacks of poor repeatability, complex data handling, and narrow linear range.
Professor XU’s team initially proposed the concept of pseudotargeted metabolomics in the year 2012. The innovation of pseudotargeted metabolomics lies in a quantitative algorithm for ion selection.
Known metabolites and unknown metabolites in samples can be quantified through the retention time locking gas chromatography-mass spectrometry-selected ions monitoring. This method combines the benefits of targeted and untargeted metabolomics techniques.
In 2013, the concept was also extended to liquid chromatography-mass spectrometry. In the following studies, XU’s research team developed additional software to automatically choose ion-pairs to enhance the efficiency of the establishment of the method.
Based on this, a pseudotargeted metabolomics method was developed to boost the number of ion-pairs included in the technique and enhance the metabolite coverage. This method is based on the sequential windowed acquisition of all theoretical fragment ions (SWATH).
In their recent work, a pseudotargeted technique was developed to further enhance the coverage. This technique is based on a two-dimensional (2D) liquid chromatography-mass spectrometry system that has a high peak capacity. With this technique, bile acids, amino acids, lysophospholipids, and carnitine can be separated and analyzed.
Such techniques have played a vital role in the analyses of diabetes, malignant tumors, and other metabolic disorders. Other laboratories have also used pseudotargeted methods and concepts.
Currently, high-coverage semi-quantitative and quantitative metabolomics techniques are gaining more attention in various fields of life sciences, and the pseudotargeted technique will play an increasingly significant role in the days to come.
Professor XU’s team has additionally optimized the development protocols of pseudotargeted metabolomics, upgraded, and offered open-accessed software and tools to enable more collaborators to make use of the pseudotargeted technique.
Source:
Journal reference:
Zheng, F., et al. (2020) Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography–mass spectrometry. Nature Protocols. doi.org/10.1038/s41596-020-0341-5.