Theoretically, high-resolution microscopy should make it possible to capture cell structures that have a resolution of a few nanometers. But in practice, this has not been achieved yet.
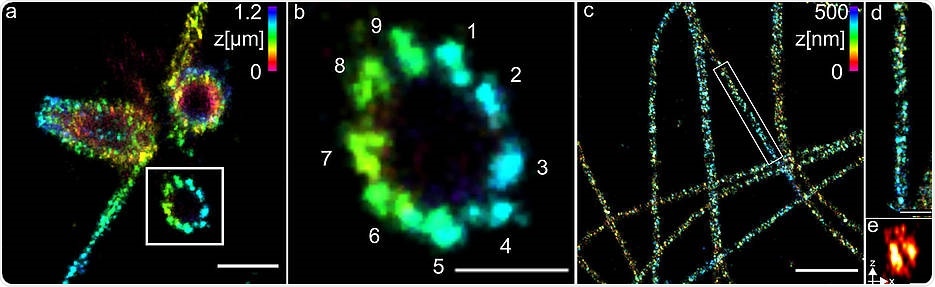
(a) Three-dimensional Ex-dSTORM of 3.2 times expanded centrioles. Measuring bar one micrometer. (b) The enlarged section of (a) shows the nine-fold symmetry of the percentile. Measuring bar 500 nm. (c) Three-dimensional Ex-dSTORM of 3.1-fold expanded tubulin filaments. Measuring bar two microns. (d) The magnification in (c) shows a tubulin filament; measuring bar 500 nm. (e) The cross-section of a tubulin filament shows its hollow structure. Measuring bar 200 nm. Image Credit: Team Markus Sauer/University of Würzburg.
The reason behind this is that antibodies that carry a fluorescent dye are often used to mark the cellular structures. Hence, the dye is not directly situated at the target structure, but rather located approximately 17.5 nm away from it. Consequently, the conceptually achievable resolution could not be realized even today, partly due to this distance error.
Publication in Nature Communications
Now, an international team of researchers has successfully overcome this barrier. The group accomplished this by integrating two super-resolution microscopy techniques—expansion microscopy (ExM) and dSTORM microscopy. The study results were published in the Nature Communications journal.
The publication was headed by a research group from the Biocenter of Julius-Maximilians-Universität (JMU) Würzburg based in Bavaria, Germany.
The team included Professor Markus Sauer, Head of the Department of Biotechnology and Biophysics, and PhD students Fabian Zwettler and Sebastian Reinhard. Professor Paul Guichard from the University of Geneva (Switzerland) and Professor Toby Bell from Monash University in Australia also contributed considerably to the study.
Obstacles to combining dSTORM and ExM
The dSTORM technique, designed by Professor Sauer’s team, was able to achieve a near molecular resolution of around 20 nm. To increase the resolution even more, a combination with expansion microscopy appeared to be promising. This combination has been available for a few years now.
In the ExM technique, the sample to be tested is first cross-linked into a swellable polymer, then the molecular interactions that occur in the sample are destroyed, and finally, the sample is allowed to swell in water. This causes an expansion—that is, the molecules that need to be imaged drift spatially apart by a factor of four.
The following reasons demonstrate why the two techniques could not be integrated until now:
- The fluorescent dyes employed for the dSTORM method to mark the molecules could not endure the polymerization of the aqueous gel.
- A buffer solution is required for the dSTORM method; however, the expanded sample reduces to its original size in such buffer solutions.
Distance error significantly reduced
By stabilizing the gel and immune staining only after expansion, we could overcome these hurdles and successfully combine the two microscopy methods.”
Markus Sauer, Professor, Department of Biotechnology and Biophysics, University of Würzburg
Consequently, the distance error melts to only 5 nm when extended by 3.2 times. For the first time, this makes it possible to perform fluorescence imaging with molecular resolution.
The scientists employed structures and centrioles that are made up of the protein tubulin to reveal the effective operation of their technique. They successfully observed tubulin tubes as hollow cylinders that have a diameter of 25 nm.
The scientists were able to capture sharp images of three groups, composed of tubulin structures at a distance of 15 to 20 nm at the centrioles. The centrioles are essentially cellular structures that significantly contribute to the cell division process.
For many important cell components, the combination of ExM and dSTORM now enables us to gain detailed insights into molecular function and architecture for the first time. The team therefore plans to apply the method to different structures, organelles and multiprotein complexes of the cell.”
Markus Sauer, Professor, Department of Biotechnology and Biophysics, University of Würzburg
Source:
Journal reference:
Zwettler, F. U., et al. (2020) Molecular resolution imaging by post-labeling expansion single-molecule localization microscopy (Ex-SMLM). Nature Communications. doi.org/10.1038/s41467-020-17086-8.