Much like humans, microbes have armed themselves with tools to identify and protect themselves against viral invaders. CRISPR-Cas serves as a key driving force of strain diversity in host-virus systems during an evolutionary fight between the host and virus.
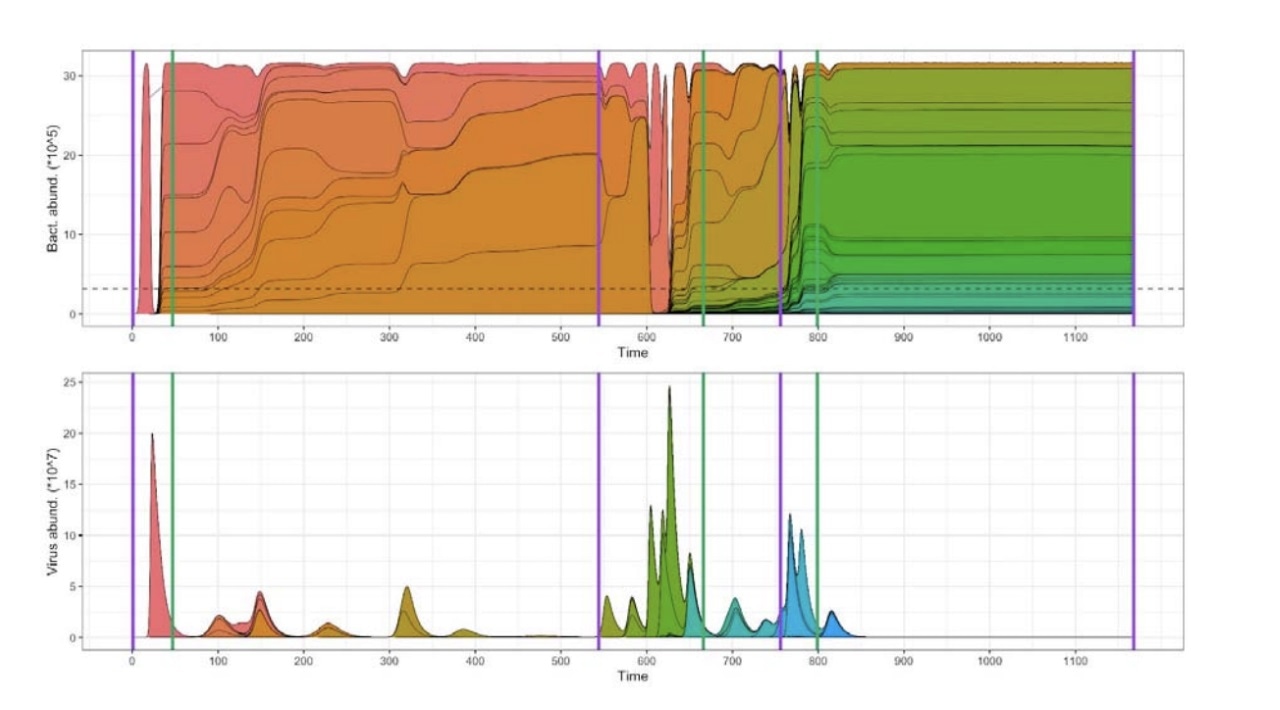
An example of typical dynamics with regime shifts. Purple lines indicate the start of VDRs while green lines indicate the start of HCRs. Top: Abundance profiles of hosts. Bottom: Abundance profiles of viruses. Image Credit: Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign.
Led by Shai Pilosof, Professor of Life Sciences at Ben-Gurion University of the Negev, Beer-Sheva, Israel; Rachel Whitaker, Professor of Microbiology at University of Illinois Urbana-Champaign; and Mercedes Pascual, Professor of Ecology and Evolution at University of Chicago, a new study points out the function of diversified immunity in arbitrating host-pathogen interactions and its eco-evolutionary dynamics.
In addition, the study involved Professor of Bioengineering and Bliss Faculty Scholar Sergei Maslov (University of Illinois Urbana-Champaign), Sergio A. Alcal´a-Corona (University of Chicago), and PhD graduate students Ted Kim and Tong Wang (University of Illinois Urbana-Champaign).
The study results have been published in the Nature Ecology & Evolution journal.
The motivation for this study was to figure out how the structure of immunity in microbial populations impacts the dynamics of virus-host interactions.”
Rachel Whitaker, Professor of Microbiology, University of Illinois Urbana-Champaign
The CRISPR-Cas system was originally developed as an adaptive immune system for microbes and is currently well known for its application in genetic engineering (Nobel Prize in Chemistry, 2020).
The system includes “protospacers”—segments of DNA from the infecting virus—integrated into the microbial host genome, named “spacers.” These spacers are used by the host molecular machinery to identify, target, and kill viruses, similar to the human adaptive immune system.
Scientists employed computational models to study the impact of microbial immune diversity on population dynamics of host-virus interactions. Their simulations showed two alternating significant regimes: the virus diversification regime (VDR) in which viruses multiply and diversify, and the host-controlled regime (HCR) in which hosts restrict virus diversification, resulting in their extinction.
When the viruses diversified in VDR regimes, so did the hosts. The viruses that could evade the host control nurtured mutations in their protospacers, thus resulting in greater encounter rates with hosts.
Hosts were capable of obtaining new spacers from these increased encounters, which led to an increase in CRISPR diversity. At the same time, the immunity network manifested weighted-nestedness, which facilitated host control.
Weighted-nestedness means that some microbial strains have redundant immunity to many viruses while others have limited immunity to a few. It is this structure that leads to the dynamics of host stability punctuated by viral epidemics.”
Rachel Whitaker, Professor of Microbiology, University of Illinois Urbana-Champaign
The team examined the weighted-nestedness immunity structure proposed by their theory by comparing the information with experimental datasets obtained from natural systems. The results showed the presence of virus control through redundant and distributed immunity in these static experimental datasets.
Whitaker added, “We next want to test this model in dynamic natural systems. We are focused on collecting high-resolution temporal data on hot springs and wastewater treatment because they are relatively simple with few viruses and microbial species.”
Scientists can better regulate microbes in industrial settings by interpreting the dynamics of host-virus populations in natural systems
Some industrial applications like wastewater treatment, yogurt, and solvent production depend on stable microbial populations. Often, these applications fail because of viral epidemics that kill these microbes. We believe that understanding CRISPRs diversity and structure can support the design of stable microbial populations that are immune to virus infection.”
Rachel Whitaker, Professor of Microbiology, University of Illinois Urbana-Champaign
Source:
Journal reference:
Pilosof, S., et al. (2020) The network structure and eco-evolutionary dynamics of CRISPR-induced immune diversification. Nature Ecology & Evolution. doi.org/10.1038/s41559-020-01312-z.