Transcription factor proteins can be described as the light switches of the human genome. These proteins bind to DNA and allow genes to turn “on” or “off.” They also initiate the significant process of duplicating DNA into an RNA template that serves as a blueprint for a novel protein.
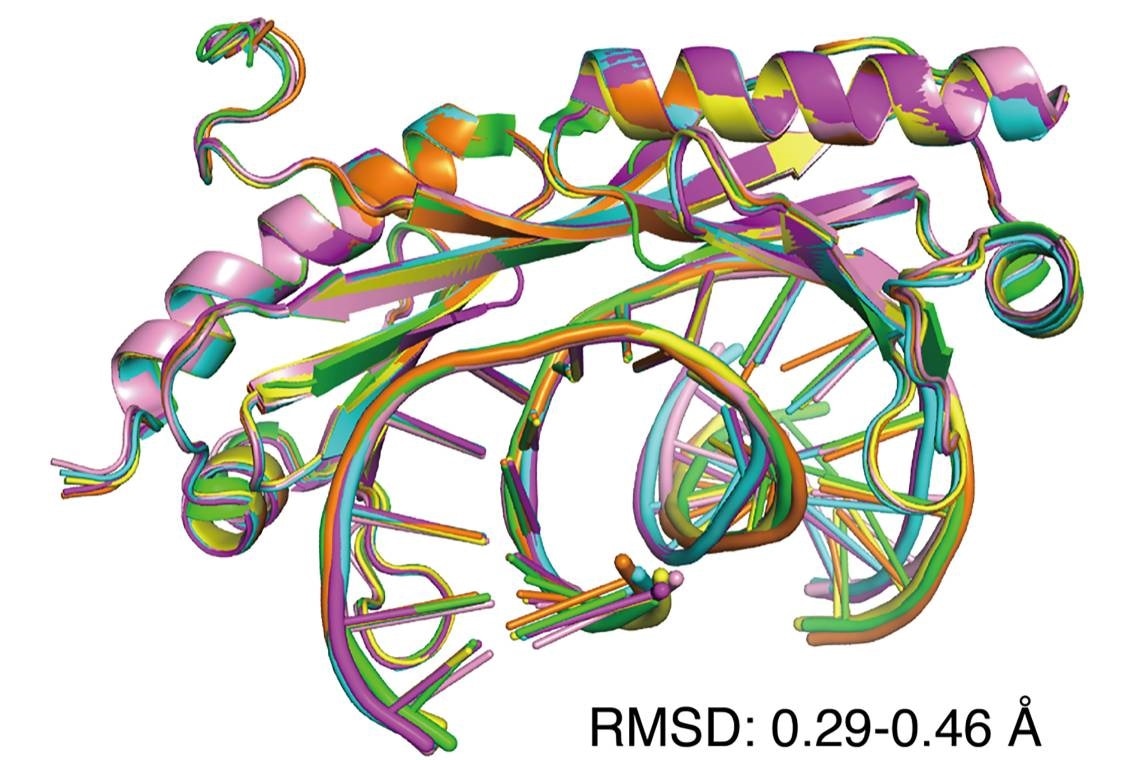
An overlay of six X-ray crystal structures of DNA shows that regular (Watson-Crick) and mismatched DNA are similarly bent by the transcription factor TBP. Mismatched DNA is preferred by the protein because it is easier to distort. Image Credit: Maria Schumacher.
The transcription factors are choosy about the type of genes they turn on, and this fact allows them to establish which rooms in the house are illuminated and which are not, or to be more precise, which elements of an individual’s genome are activated.
Now, a group of scientists from Duke University has discovered that transcription factors tend to attach robustly to “mismatched” parts of DNA, that is, sections of the code that were not copied properly. The robust binding of transcription factors to mismatched regions of regulatory DNA could be a means in which haphazard mutations become an issue that results in numerous diseases, like cancer.
The study results were published in the Nature journal on October 21st, 2020
Generally, the replication of DNA in the body occurs smoothly with nucleotides coupling with their complementary base pair and collectively marching via the cycle in an intended A-T and C-G manner. But as Gordan explained it, “no polymerase is perfect” and from time to time, a nucleotide will be coupled with the wrong partner leading to a mismatch.
A team of researchers, headed by Raluca Gordan PhD—a computational biologist from Duke University—pipetted transcription factor proteins on slides that were pre-blotted with scores of DNA molecule samples. They demonstrated that the proteins shared a stronger bond with the parts of DNA containing the mismatched base pairs than with those containing ideally matched base pairs or a “normal” structure of DNA.
But scientists are clueless as to what makes these “mistakes” an attractive site of binding for the transcription factor proteins. To gain a deeper understanding, Gordan—an associate professor in the Department of Biostatistics and Bioinformatics and the Department of Computer Science—collaborated with Hashim Al-Hashimi, PhD, a James B. Duke Professor of Biochemistry. Hashimi is also an expert in DNA dynamics and structure who works just across the street.
He studies nucleic acids (RNA and DNA) and their interactions with small molecules and proteins, with the notion that how these biomolecules appear and move is as significant for their function as their chemical characteristics.
Observing the experimental results, both Gordan and Al-Hashimi ultimately concluded that the strong interaction between mismatched DNA and transcription factor proteins has plenty to do with laziness. Upon binding to DNA, a transcription factor protein should spend energy to deform the site, for instance, by bending the DNA to its will. But mismatched parts of DNA are already deformed, and hence the transcription factor protein has to perform less work.
Gordan added, “That’s when the transcription factor doesn’t need to pay that energetic penalty to get the job done.”
If we are ever to attain a deep and predictive understanding of how DNA is recognized by proteins in cells, we need to go beyond the conventional description in terms of static structures and move towards describing both DNA and the protein molecules that bind to them in terms of dynamic structures that have different preferences to adopt a wide range of shapes.”
Hashim Al-Hashimi, PhD, James B. Duke Professor of Biochemistry, Duke University
Gordan added that in the future, the researchers hope to figure out how this interaction is associated with the development of the disease. If a mismatched base pair, attached strongly by a transcription factor, passes the DNA replication cycle without being corrected by another form of protein—called a repair enzyme—it can become a mutation, and such mutations can cause genetic diseases, for example, neurodegeneration and cancer.
We are now convinced that the interactions between transcription factors and mismatches are really strong. So the next step is to understand what this means for the cell. We already know that regulatory regions of the genome harbor more cancer mutations than expected by chance. We just do not know why.”
Raluca Gordan, PhD, Computational Biologist, Duke University
“The strong interactions between transcription factors and DNA mismatches, which could interfere with repair of the mismatches, provide a novel mechanism for the accumulation of mutations in regulatory DNA,” Gordan concluded.
Source:
Journal reference:
Afek, A., et al. (2020) DNA mismatches reveal conformational penalties in protein–DNA recognition. Nature. doi.org/10.1038/s41586-020-2843-2.