Several drugs have been widely employed to combat SARS-CoV-2 infection, including dexamethasone and colchicine to suppress excessive immune response; antiviral drugs remdesivir, favipiravir, and ribavirin; and antimalarial drugs chloroquine phosphate and hydroxychloroquine for combined suppression of viral replication and immune response.
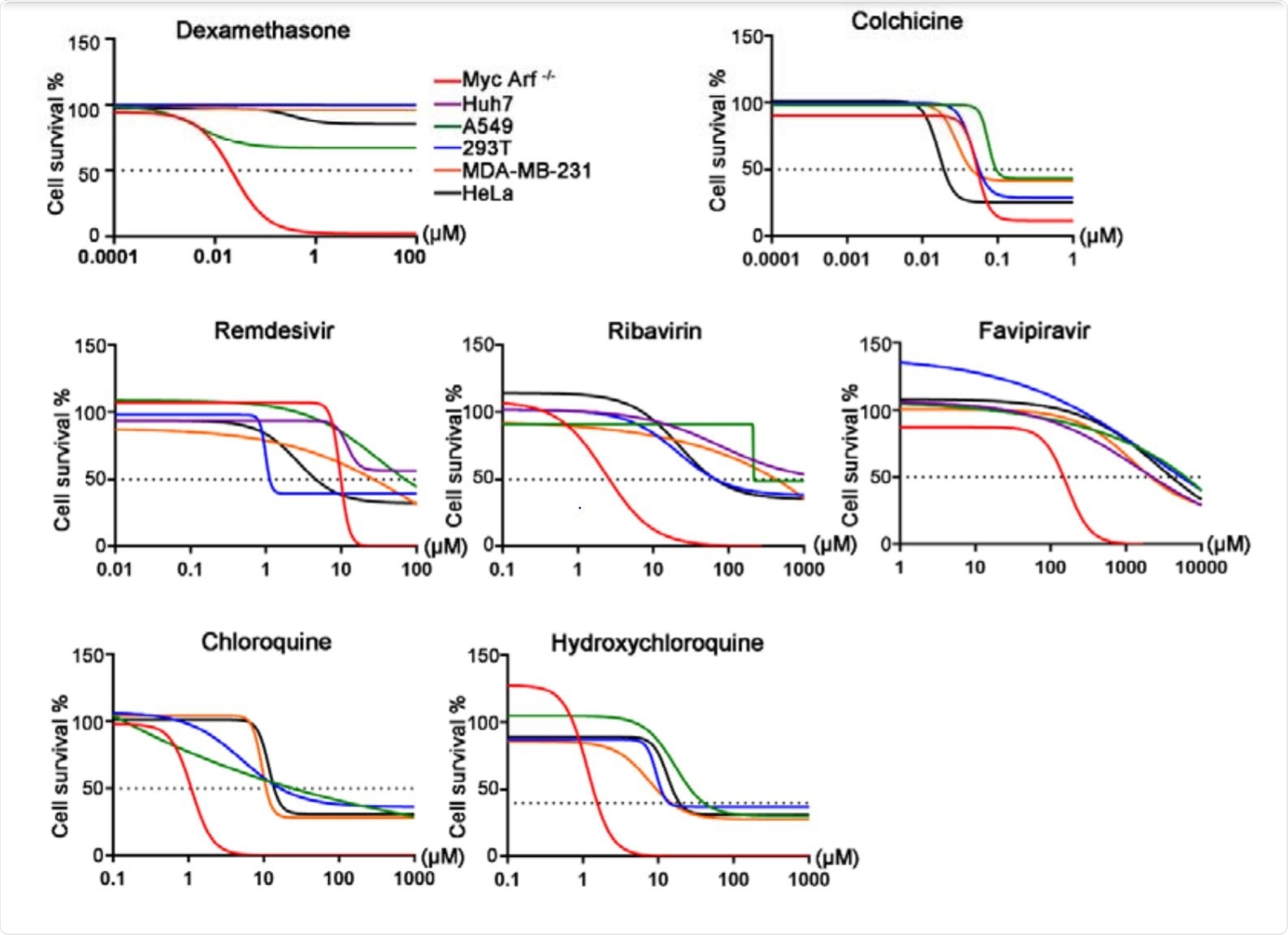
Image Credit: https://www.biorxiv.org/content/10.1101/2020.10.26.356279v1.full.pdf

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
However, little is currently known with regards to genetic biomarkers that may affect the efficacy and toxicity of these drugs. A paper recently uploaded to the preprint server bioRxiv* by Jiang et al. (October 2020) aimed to utilize genome-wide CRISPR screening to identify genes involved in modulating the activity of these drugs.
A cas9-expressing Eμ-Myc; Arf-/- cell line was selected for its sensitivity to drug-induced apoptosis, as many of the drugs tested are not particularly cytotoxic and so genetic predispositions to drug-induced damage are hard to identify in more robust cell lines.
A retroviral backbone was used to introduce a genome-wide sgRNA library into the cells, and the cells were then exposed to the drugs at a concentration that allowed 10-20% cell viability. Determining the sgRNAs that were then highly represented in the surviving cells allowed the group to identify the genes involved in modulating the efficacy of these drugs.
Which genes affect the efficacy of each drug?
Colchicine is an immunomodulatory agent that acts by destabilizing microtubules and mitotic spindles in immune cells, alleviating the symptoms of SARS-CoV-2 infection associated with an over-active immune response. sgRNAs associated with microtubule depolymerizing protein KIF2C were highly expressed in the surviving cells, demonstrating the validity of the system.
Among the most highly expressed were TMEM161B and POU2F1, genes associated with resistance to other microtubule destabilizing drugs. If a patient suffers from deficiencies in these genes, colchicine may be unable to sufficiently suppress the immune response.
Dexamethasone is a glucocorticoid used to suppress lymphocyte proliferation in many autoimmune diseases. sgRNAs associated with transcription factor NR3C1, the glucocorticoid receptor for dexamethasone, were greatly enriched in surviving cells. PTGES3 and TSC22D3 were also upregulated, which are a chaperone protein for NR3C1 and an immunosuppressive protein induced by glucocorticoids, respectively. The inhibitor of the DNA Binding 3 (ID3) gene, which plays a role in inhibiting the transcriptional activity of NR3C1, was simultaneously found to be down-regulated.
Ribavirin is a broad-spectrum antiviral drug that is converted into a nucleotide triphosphate (NTP) analog, allowing it to inhibit RNA-dependent RNA polymerase reactions. Three enzymes involved in catalyzing the formation of NTP analogs were noted to be highly expressed in surviving cells: adenosine kinase, adenylosuccinate synthase, and NUDT2.
Remdesivir is also an antiviral drug that degrades to an adenosine monophosphate analog within the cell, involving breaking a phosphoramidate bond, before being converted into an ATP analog that interferes with RNA-dependent RNA polymerase. Surviving cells exhibited upregulated SLC29A3, a transporter protein found on the membrane of cells that appears to be highly specific towards remdesivir, and HINT1, an enzyme that cleaves phosphoramidate bonds.
Favipiravir is an antiviral prodrug, and therefore requires extensive metabolic conversion in vivo before efficaciously entering the target cell. In this study, only cell death genes were noted to be significantly enriched in the surviving population, while some drug inactivation mechanisms were depleted.
With regards to drug transport and inactivation mechanisms, the group noted that surviving cells often had upregulated drug efflux pumps that were specific to the drug of interest. Remdesivir was best removed from cells expressing ABCC1, while colchicine was most effectively removed by ABCC1 and ABCB1. High expression of nucleotidase gene NT5C2 was also noted to significantly reduce the cytotoxicity of both remdesivir and favipiravir, inactivating them.
As a confirmatory test Huh7 cells with knocked-out genes were infected with the mechanistically similar Zika virus and exposed to the drugs individually. The antiviral activity of remdesivir was 30-100 times lower in the SLC29A3 and HINT1 knockout cells, while the effects of the drug were enhanced in the ABCC1 or NT5C2 knockout cells. Ribavirin was similarly less efficacious in ADK, ADSS, and NUDT2 knockout cells. TMEM161D, RQCD1, PTBP1, FLCN, and FNIP knockout cells were far less sensitive to colchicine-induced death, while ID3 knockout promoted the cytotoxic effects of dexamethasone.
This work will ideally open the door to improved personalized medicine experiences for COVID-19 patients, ensuring that they are best matched to the ideal drug. For example, these findings suggest that NT5C2 is involved in the dephosphorylation of NMP analog for remdesivir and favipiravir, but not ribavirin.
Therefore, patients with a defective NT5C2 gene are likely to show variation in the efficacy of remdesivir and favipiravir compared to treatment with ribavirin. The authors note that combinatorial treatments with multiple drugs may be of benefit once an individual’s genetic profile is assessed.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
https://www.biorxiv.org/content/10.1101/2020.10.26.356279v1.full.pdf