The structural proteins of SARS-CoV-2 play a key role in self-arrangement and particle formation, and current work suggests that an assembly is formed in the lipid bilayer of the endoplasmic reticulum, aided by interactions between the host membrane and the spike, envelope, and nucleocapsid proteins of the virus.
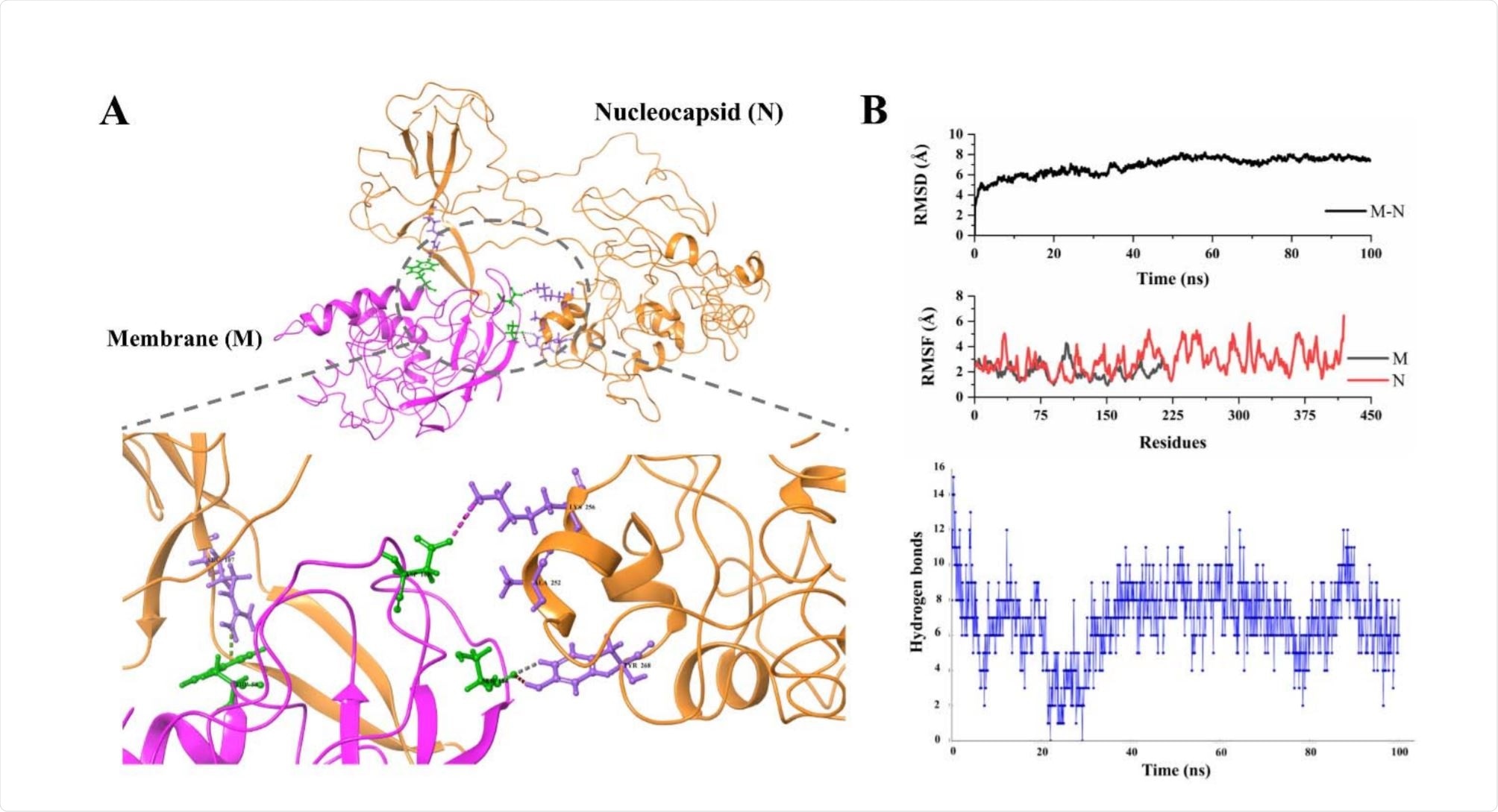
Image Credit: https://www.biorxiv.org/content/10.1101/2020.10.30.363002v1.full.pdf

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
A computational study of these interactions was performed by Kumar et al. (2020) in a paper recently uploaded to the pre-print server bioRxiv* (November 2020), examining the interactions between these proteins and a host membrane protein.
Protein-protein interactions are the basis on which a virus can replicate and commandeer the host’s cellular machinery, and thus a greater understanding of these interactions is advantageous in identifying druggable targets and the pathogenic mechanism of action. The spike protein (S) of SARS-CoV-2 consists of two subunits that have been shown in other work to recognize the human receptor ACE2, mediating the fusion of the host and viral membranes.
The function of nucleocapsid proteins (N) is usually in packaging viral RNA, and in the case of SARS-CoV-2 contains the N- and C-terminal RNA-binding domains, with serine and arginine-rich domain linking them. The envelope protein (E) is a transmembrane protein with an external N-terminal domain and an internal C-terminal domain. The group constructed the S, N, and E proteins based on structural analytical methods such as X-ray crystallography and cryogenic electron microscopy, and performed protein-protein docking experiments towards a host membrane protein (M) to observe the binding and stability.
Envelope-Membrane interactions
Both M and E proteins contain a number of aromatic hydrogen bonds and pi-pi stacking, binding together relatively strongly with a binding energy of -10.1 kcal/mol once docked. The interaction was noted to be largely via 5 hydrogen bonds on the CYS33, PHE37, TYR39, and HIS125 residues of the M protein to the PHE26, PHE23, TYR59, SER60, and LUE27 residues of the E protein.
Overall the bond was relatively stable with a root-mean-square deviation (RMSD) of around 6 Å, up to 9 Å after 100 ns.
Spike-Membrane interactions
Multiple hydrogen bonds, pi-cation, and pi-pi stacking was observed between the S and M proteins, with strong binding energy of -18.4 kcal/mol in the docked complex. Throughout the course of a 100 ns simulation, an average of 16 hydrogen bonds were formed, while the RMSD started at only 5 Å and stabilized at 15-20 Å after around 20 ns.
Much of the observed movement was due to the loosely packed S protein, though the C-terminal surface of interaction was noted to be stable with an RMSD of around 8 Å.
Nucleocapsid-Membrane interactions
Three amino acid residues, in particular, were noted to interact on each protein, with Arg107, Lys256, and Tyr268 residues of the N protein interacting with Trp58, Asp163, and Ser184 of the M protein, by Pi-cation, hydrogen bonding, and aromatic/hydrogen bonding, respectively. There was an average of 7 hydrogen bonds between the proteins over the course of the 100 ns simulation.
A weaker docked binding energy of -8.3 kcal/mol was observed, though the complex was found to be highly stable, with an average RMSD of only 6.8 Å, however, non-interacting regions of the N protein fluctuate greatly.
Conclusions
This work demonstrates the interactions between host M proteins and the structural S, N, and E proteins of SARS-CoV-2, aiding in proper virus assembly.
The M protein has been shown to be essential in this function, with the entirety of the C-terminus ectodomain interacting with the N protein, and multiple distinct regions interacting with either the S or E proteins. This reinforces previous reports of particle assembly taking place largely in the endoplasmic reticulum-Golgi intermediate compartment, being released by exocytosis.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
https://www.biorxiv.org/content/10.1101/2020.10.30.363002v1.full.pdf