Scientists from the University of Illinois Urbana Champaign have designed a new method that uses a combination of artificial intelligence and label-free imaging to observe unlabeled live cells across an extended time.
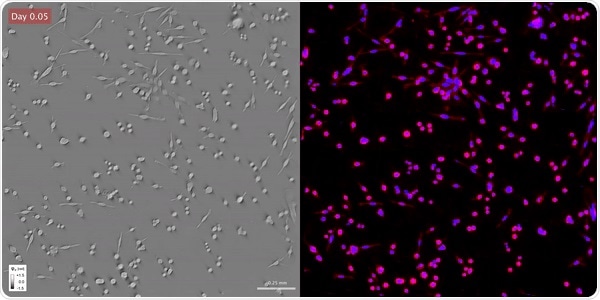
Time-lapse gradient light interference microscopy, or GLIM, left, and phase imaging with computational specificity imaged over seven days. Image Credit: Beckman Institute for Advanced Science and Technology.
This method offers promising applications for investigating the viability and pathology of cells.
The study titled, “Phase imaging with computational specificity (PICS) for measuring dry mass changes in subcellular compartments,” was published in the Nature Communications journal.
Our lab specializes in label-free imaging, which allows us to visualize cells without using toxic chemicals. However, we cannot measure specific attributes of the cell without using toxic fluorescent dyes. We have solved that problem in this study.”
Gabriel Popescu, Professor of Electrical and Computer Engineering and Director of Quantitative Light Imaging Laboratory, Beckman Institute for Advanced Science and Technology
According to Mikhail Kandel, a graduate student in Popescu’s group, “We had this idea that computational methods could estimate what the sample would look like without actually killing the cells.”
Using their non-destructive label-free method, the investigators initially imaged the cells over many days. Toward the end of the experiment, the team stained the samples and applied deep learning—a subset of machine learning—to find out where the fluorescence dyes would be actually placed.
Kandel added, “This lets us estimate the stain in our initial movies without actually staining the cells.”
“Although AI has been used in the past to create one type of imaging from a different type of staining, we were able to program it to analyze the images in real-time. Using deep learning, we were able to look at cells that had never been tagged with any dye, and the algorithm was able to precisely locate different parts of the cell,” Popescu added.
Another advantage of the technique is that we can perform experiments over the span of many days. The cells remain alive even after more than a week. This cannot be done with fluorescent dyes since the chemical toxicity might kill the cells.”
Yuchen He, Graduate Student, Beckman Institute for Advanced Science and Technology
Yuchen He is part of Popescu’s group.
Kandel further added, “This study highlighted the potential of AI-based techniques to learn complicated models such as the concentration of specific dyes, which goes beyond the capabilities of the naked eye. The more we can teach our method to recognize patterns, the more kinds of experiments can be performed without resorting to killing the cells.”
Currently, the team is attempting to adapt deep learning algorithms across various biological samples and cell lines.
Training deep learning models require a large amount of data because we want to ensure that they work well in different scenarios. Fortunately, our imaging instruments make it easy for us to generate the needed training data in an efficient fashion.”
Mikhail Kandel, Graduate Student, Beckman Institute for Advanced Science and Technology
“These deep learning algorithms can be used for several applications. We can assess the cell viability over a long time without labeling the cells, we can differentiate between different cell types in diseases, and we can study different cellular processes,” Popescu concluded.
Source:
Journal reference:
Kandel, M. E., et al. (2020) Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments. Nature Communications. doi.org/10.1038/s41467-020-20062-x.