Polyacrylamide gel electrophoresis is a technique that allows high-resolution isolation of proteins separated from biological specimens. But this technique needs more than a single day of pretreatment to retrieve the isolated proteins confined within the gel for detection through mass spectrometry.
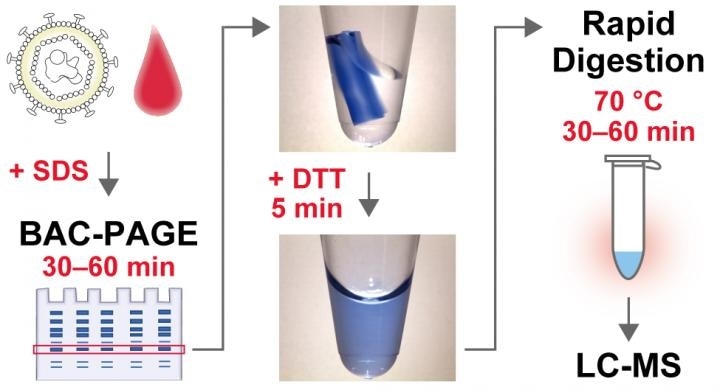
Dissolvable BAC cross-linked gels allow rapid and lossless recovery of protein biomarkers separated by SDS-polyacrylamide gel electrophoresis and facilitate analysis by mass spectrometry. Image Credit: Reprinted with permission from Journal of Proteome Research 2020 American Chemical Society (ACS).
BAC-DROP is a unique electrophoresis method that utilizes a dissolving form of polyacrylamide gel that helps complete sample pretreatment in approximately five hours. The advanced technology will allow viruses and disease protein markers to be diagnosed quickly.
Mass spectrometry (MS) is a robust technique developed for biomarker studies because it allows for precise and extremely sensitive measurement of molecules of interest in clinical specimens. The use of MS technique in clinical diagnosis, like neonatal metabolic screening, has been advancing with an emphasis on metabolite markers.
At present, MS measurement of proteins is predominantly used for innovative marker discovery studies; however, it is increasingly applied in clinical marker diagnosis as an alternative to immunoassays.
MS-based measurement of protein biomarkers is chiefly conducted by a bottom-up method using peptide fragments acquired by enzymatic digestion of proteins with trypsin. Conventional digestion procedures need a reaction time of over 20 hours, which is a rate-limiting factor in specimen preparation workflows.
Even though MS-based protein measurement is extremely sensitive, serum proteome and plasma are highly complex, and disruption by other components presents a major problem.
For highly accurate identification of target markers, techniques for the pre-removal of key serum protein components, including albumin, or selective enrichment of markers of interest using antibody columns have been already reported; however, the off-target impact on quantitative outcomes and the challenge of processing several specimens still poses a difficulty.
In the new analysis, the researchers focused on dissolving polyacrylamide gels using N,N'-Bis(acryloyl) cystamine (BAC) as a cross-linker to address these issues. BAC cross-linked polyacrylamide gels rapidly dissolve by reduction treatment, making it possible to retrieve proteins that have leaked into the solution.
The researchers observed that the recovered proteins were appropriate for rapid digestion of trypsin under extreme temperature conditions and they succeeded in defining a high-throughput sample preparation technique for MS-based quantification of biomarkers. This method was called BAC-Gel Dissolution to Digest PAGE-Resolved Objective Proteins, or BAC-DROP for short.
High-resolution proteome fractionation using the BAC-DROP method is specifically effective for MS-based quantification of specified trace marker proteins extracted from clinical specimens.
When BAC-DROP is introduced into the MS-based measurement workflow of the inflammatory biomarker C-reactive protein (CRP), the team was able to pretreat the samples and completed this task in just five hours. Moreover, they were able to efficiently measure the CRP from a human serum sample of 0.5 μL.
The researchers succeeded in combining serological diagnosis of hepatitis B virus (HBV) infection by HBsAg quantification with both MS and BAC-DROP. In the recent past, interest in MS diagnosis of viral infections has been quickly increasing, as demonstrated by COVID-19 diagnosis.
The BAC-DROP-based high-throughput sample preparation method, demonstrated in this analysis, would be relevant not only to HBV but also to other infectious viral disease specimens.
Source:
Journal reference:
Takemori, A., et al. (2020) BAC-DROP: Rapid Digestion of Proteome Fractionated via Dissolvable Polyacrylamide Gel Electrophoresis and Its Application to Bottom-Up Proteomics Workflow. Journal of Proteome Research. doi.org/10.1021/acs.jproteome.0c00749.