A fine balance between the production and degradation of biomolecules is required for optimal cell function. Autophagy is a mechanism through which cells break down and recycle their own components. This helps to clean up and preserve the internal environment of the cell and thus ensure the smooth working of cellular processes.
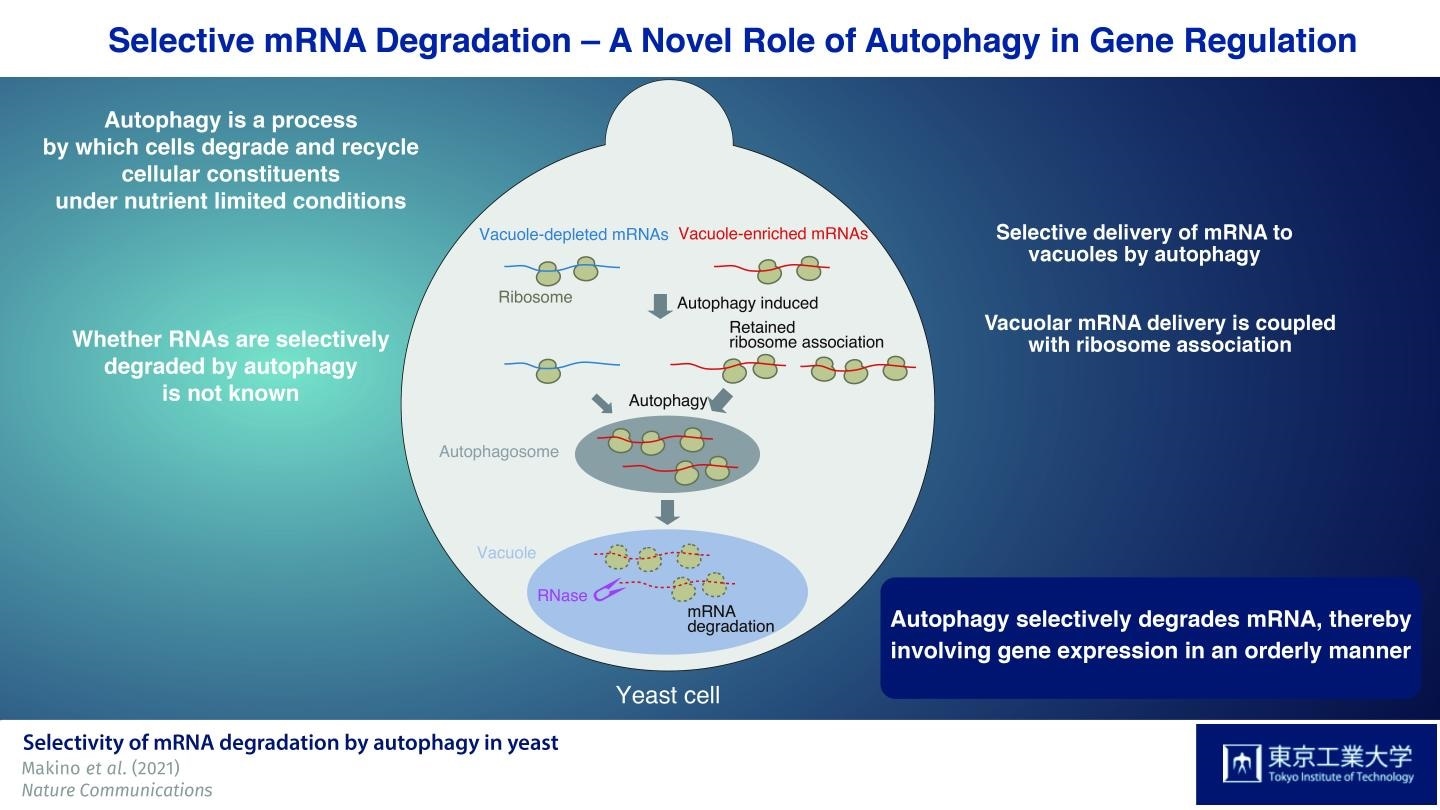
A subset of mRNAs is preferentially delivered to vacuoles by autophagy. This mRNA delivery is coupled to mRNA translation. The persistence of ribosome association with mRNA enhances selective mRNA delivery to vacuoles. mRNAs delivered to the vacuole by autophagy are subsequently degraded by the nuclease. Image Credit: Tokyo Institute of Technology.
When cells are exposed to stresses, such as nutrient deprivation, autophagy is induced strongly. This process acts under these conditions to deliver nutrients by breaking down unneeded cellular content. Autophagy substrates are supplied to lysosomes in mammals or vacuoles in yeast for degradation by “autophagosomes,” which are double-membrane vesicles.
Although autophagy was initially thought to be a non-selective mechanism that randomly separates substrates in the cell’s cytoplasm, studies have demonstrated that specific cellular components, including a subset of proteins and superfluous or impaired cell organelles, are separated in a selective manner.
Unlike this well-established targeting of proteins and organelles by autophagy, the question of whether RNAs are exposed to autophagy and whether they are selectively destroyed is yet to be answered.
In their new study recently published in the Nature Communications journal, researchers from the Tokyo Institute of Technology (Tokyo Tech) and RIKEN performed a comprehensive analysis of the preferred degradation through autophagy of messenger RNAs (mRNAs). mRNAs contain the required data to produce cellular protein and bind ribosomes for the production of proteins.
The team’s results were explained by Professor Yoshinori Ohsumi from Tokyo Tech. He is also the corresponding author of the study and had received the 2016 Nobel Prize in Physiology or Medicine for his groundbreaking research work in the field of autophagy.
We have previously shown that RNA delivered to the vacuole via autophagy in yeast cells, where it is degraded by vacuolar nucleases. The question of whether RNA degradation by autophagy occurs preferentially, however, remains unaddressed. This difficult to address question was the starting point of this project.”
Yoshinori Ohsumi, Study Corresponding Author and Professor, Tokyo Institute of Technology
Since RNAs that build up in the vacuole are enzymatically decomposed by the nuclease Rny1, the researchers initially created a yeast strain without this enzyme. With the help of this strain, they successfully separated and detected RNAs that built up in the vacuole.
The researchers then used the autophagy-inducing drug rapamycin to examine the special features of mRNA species supplied to the vacuole in Rny1-deficient cells when autophagy is triggered. Most significantly, they discovered that autophagy-mediated delivery of mRNAs to vacuoles is not random but selective in nature.
The team then identified “'vacuole-depleted” and “vacuole-enriched” mRNAs by characterizing the different species of mRNAs and performing a wide analysis of the types of mRNAs found in these cells. Fascinatingly, housekeeping mRNAs, like those encoding proteins that play a role in the biosynthesis of amino acids, are probably supplied to vacuoles. On the other hand, mRNAs needed for producing proteins with regulatory functions, like protein kinases, were mainly detected in the vacuole-depleted mRNA fraction.
The researchers also showed that mRNAs that undergo translation are transmitted to the vacuole, indicating that this is a translation-dependent process. Furthermore, persistent mRNA-ribosome association after rapamycin therapy was found to be a major determinant of vacuolar mRNA delivery at the time of autophagy-mediated degradation.
Dr Makino and Professor Ohsumi emphasized the significance of autophagy in gene regulation, and stated, “Our findings suggest that autophagy regulates mRNA degradation at the translation step, thereby enabling a rapid and sensitive switch from ribosome-associated mRNAs to expression of mRNAs that are essential for an effective response to stress. Preferential degradation of ribosome-mRNAs by autophagy is therefore very likely to determine the fate of individual mRNAs as cells adapt to new conditions.”
Source:
Journal reference:
Makino, S., et al. (2021) Selectivity of mRNA degradation by autophagy in yeast. Nature Communications. doi.org/10.1038/s41467-021-22574-6.