Researchers from Sanford Burnham Prebys have discovered, at the atomic level, how a portion of a protein known as PLEKHA7 communicates with a cell’s membrane to control crucial intercellular communications.
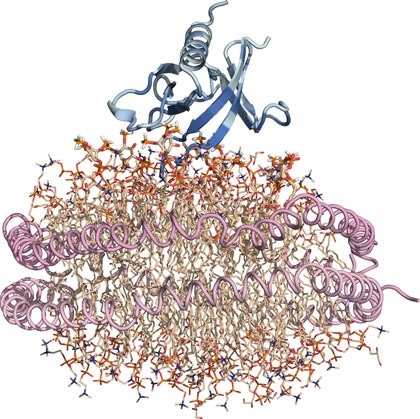
Nanodisc “substitute” cell membrane: Protein is blue, lipids are wheat, and the two helical lipoproteins that corral the lipid nanodisc and hold it together are pink. Image Credit: Sanford Burnham Prebys Medical Discovery Institute.
The findings, reported in the Structure journal, indicate that hotspots inside the PLEKHA7 protein could be potential drug targets. Such targets may be crucial in the development of new therapies for advanced breast, ovarian, and colon cancers.
Pleckstrin homology (PH), the domain or region in the PLEKHA7 protein examined by the researchers, is generally found in proteins that control cell movement as well as other crucial cellular activities. Diseases like cancer can occur if there is a disruption in the communication between the PH domain and the lipids comprising cell membranes.
PH domains have been studied for some time but examining their interactions with membrane-associated lipids has been challenging. We optimized our structural investigations by using an artificial membrane disc several nanometers in size. This nanodisc is a ‘substitute’ cell membrane and has been an important tool in overcoming obstacles in studying structural aspects of fatty interfaces, such as those involving lipids.”
Francesca M. Marassi, PhD, Professor and Director, Cell and Molecular Biology of Cancer Program, Sanford Burnham Prebys Medical Discovery Institute
Marassi is also the corresponding author of the study.
The researchers used a variety of methods to pinpoint the exact locations where interactions occurred between the PH domain and lipids in a cell’s membrane.
All three methods were equally significant: X-ray crystallography offered a glimpse of the structure; magnetic resonance revealed the exact relationship between the lipid membrane surface and the PH domain, and computer simulations combined all of this data to create a dynamic movie of this interaction.
We were able to determine that the PH domain interacts with the cellular membrane at several locations simultaneously, thereby demonstrating the key role of the membrane as a platform for directing cell signaling and adhesion. We found at least three sites along the PH domain that are engaged in binding lipids.”
Francesca M. Marassi, PhD, Professor and Director, Cell and Molecular Biology of Cancer Program, Sanford Burnham Prebys Medical Discovery Institute
“Having multiple sites is crucial because if you think of the PH binding sites as a zipper between proteins and lipids, each additional notch engaged by a zipper makes the binding stronger and more resistant to being pulled apart,” added Marassi.
One of the reasons the researchers were keen on studying the PLEKHA7 protein was that it has recently been recognized as a promising anti-cancer target. But the molecular mechanism of action has been vague.
The team was also interested in the PH domain because it has been linked to advanced renal, ovarian, and breast cancers and, most importantly, plays a key role in colorectal cancer.
Clinicians have shown that PLEKHA7 is elevated in patients with colorectal cancer, and its levels increase as the disease worsens. It has also been shown that inhibiting PLEKHA7 decreases cellular proliferation and migration. Therefore, the PLEKHA7 PH domain might be a useful drug target, but more work is needed to develop agents with greater potency and optimized pharmacological properties.”
Francesca M. Marassi, PhD, Professor and Director, Cell and Molecular Biology of Cancer Program, Sanford Burnham Prebys Medical Discovery Institute
As a subsequent step, the team has planned to share their structural images with collaborators in other laboratories at Sanford Burnham Prebys and elsewhere to test drugs that target the PH domain as well as lipid membrane binding locations.
Source:
Journal reference:
Aleshin, A. E., et al. (2021) Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids. Structure. doi.org/10.1016/j.str.2021.03.018.