Mitigating and controlling the impact of ice growth is critical for infrastructure safety, preserving frozen cells, and improving the texture of frozen foods.
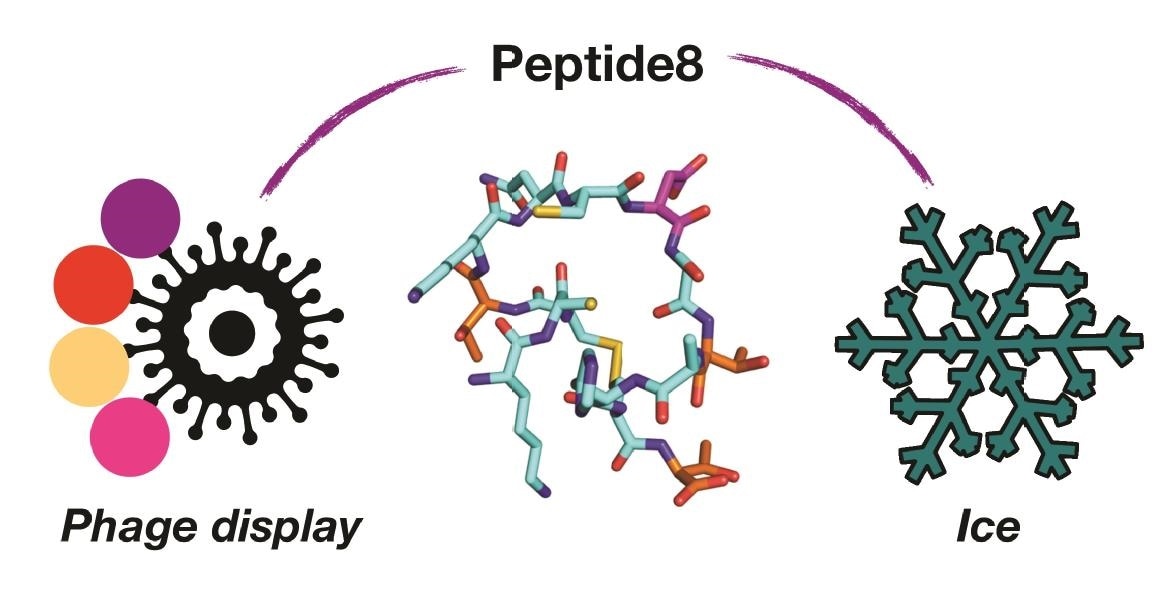
Using viruses (phage display) to identify the one molecule in a billion (peptide8) that controls the formation of ice. Image Credit: University of Warwick.
A phage display platform was used by Warwick Scientists in collaboration with Swiss researchers to discover new, tiny peptides that act like larger antifreeze proteins. This opens the door to new, easier-to-make cryoprotectants.
Ice binding proteins, which include antifreeze proteins, are developed by a wide variety of species, ranging from fish to insects to plants, to avoid ice damage. Also, in the presence of a huge amount of water (which ice is the solid form of), the proteins can recognize and attach to ice.
New antifreeze proteins have typically been discovered by isolation from the organisms. The team took a quite different method in this work, scanning billions of potential peptides to identify those that could bind to ice. This was accomplished by Phage Display, a technology in which a virus is used to produce a large number of peptides, and those that “bind” to the ice can be isolated.
Using this, a cyclic peptide with 14 amino acids (very short relative to a typical protein) that could bind to ice was found. The researchers used computer simulations to figure out how the peptide binds to the ice, which is impossible to do with only “wet” laboratory techniques.
The researchers have demonstrated how this short peptide can be used to aid in the purification of other proteins using ace affinity purification.
By finding these short peptides, it means a research team can now easily create (or buy) modified peptides to learn and probe how these bind with ice, allowing them to design new cryoprotectants with simpler structures and hence lower cost.
In the study “A Minimalistic Cyclic Ice-Binding Peptide from Phage Display,” published in Nature Communications, an international group including the University of Warwick and led by EPFL, Switzerland, showed the use of phage display to explore new minimalistic antifreeze peptides that traditional methods could not accomplish. This would not enable billions of potential structures to be monitored.
This work highlights that even very small changes within the structure of these peptides can make a huge difference in their ability to control the formation of ice. Our computer simulations allowed us to identify and understand the importance of these structural changes—which is a key step toward the rational design of synthetic cryoprotectants.
Dr Gabriele Sosso, Assistant Professor, Department of Chemistry, University of Warwick
“It is such a privilege to be able to leverage both the experimental work of Gibson’s group and the computational resources of the SCRTP. Truly, Warwick is a great place to be if you want to understand how ice forms and what can we do to have a say in this process,” added Sosso.
We have been working on developing synthetic tools to understand, and interfere with, ice growth processes with an aim of helping develop new cryoprotectants. This work was really exciting, as we made use of biotechnology tools (phage) to discover small, cyclic, peptides which are remarkably potent.
Matthew Gibson, Professor, Department of Chemistry and Warwick Medical School, University of Warwick
“These peptides are easy to synthesis and modify and will accelerate our research in this field. It also highlights the growing ‘team ice’ collaborative network at Warwick, combining experimental and computation studies together. We are also grateful for the support from the IAS at Warwick, which allowed Dr. Stevens to visit us to complete this work, showing the need to support international scientific collaborations,” concluded Gibson.
Source:
Journal reference:
Stevens, C. A., et al. (2021) A minimalistic cyclic ice-binding peptide from phage display. Nature Communications. doi.org/10.1038/s41467-021-22883-w.