Antibiotics used to treat common bacterial infections such as sinusitis and pneumonia, according to scientists at the University of Illinois Chicago, may potentially be used to treat human illnesses such as cancer, at least theoretically.
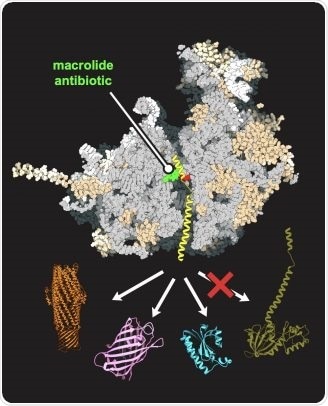
An antibiotic (green), bound in the human-like yeast ribosome (gray), allows for the synthesis of some proteins (represented in orange, purple, and blue) but not others (dark green). Image Credit: Maxim Svetlov/University of Illinois Chicago.
The UIC College of Pharmacy team demonstrated in laboratory tests that eukaryotic ribosomes may be changed to respond to antibiotics in the same manner as prokaryotic ribosomes do, as described in a recent Nature Communications research.
Plants, fungi, and animals, like humans, are eukaryotes; they are composed of cells with a well-defined nucleus. By contrast, bacteria are prokaryotes. They are built out of cells that lack a nucleus and have a unique size, properties, and structure.
In eukaryotic and prokaryotic cells, the ribosomes, which are responsible for protein synthesis required for cell development and reproduction, differ as well.
Some antibiotics, used for treating bacterial infections, work in an interesting way. They bind to the ribosome of bacterial cells and very selectively inhibit protein synthesis. Some proteins are allowed to be made, but others are not. Without these proteins being made, bacteria die.”
Alexander Mankin, Study Senior Author and Alexander Neyfakh Professor of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, University of Illinois Chicago
When antibiotics are used to treat an infection, the patient’s cells remain unaffected because the medications are not designed to attach to the differently shaped ribosomes of eukaryotic cells.
Because there are many human diseases caused by the expression of unwanted proteins — this is common in many types of cancer or neurodegenerative diseases, for example — we wanted to know if it would be possible to use an antibiotic to stop a human cell from making the unwanted proteins, and only the unwanted proteins.”
Alexander Mankin, Study Senior Author and Alexander Neyfakh Professor, Medicinal Chemistry and Pharmacognosy, College of Pharmacy, University of Illinois Chicago
Mankin and study first author Maxim Svetlov, research assistant professor in the department of pharmaceutical sciences, turned to yeast, a eukaryote with cells identical to human cells, to find a solution.
The research team, which included partners from Germany and Switzerland, performed a “cool trick”. We engineered the yeast ribosome to be more bacteria-like.”
Alexander Mankin, Study Senior Author and Alexander Neyfakh Professor, Medicinal Chemistry and Pharmacognosy, College of Pharmacy, University of Illinois Chicago
Mankin and Svetlov’s group utilized biochemistry and fine genetics to modify one of over 7,000 nucleotides in yeast ribosomal RNA, which was enough to allow a macrolide antibiotic—a popular family of antibiotics that function by binding to bacterial ribosomes—function on the yeast ribosome.
The researchers used this yeast model to do genomic profiling and high-resolution structural analysis to learn how each protein in the cell is generated and how the macrolide interacts with the yeast ribosome.
“Through this analysis, we understood that depending on a protein’s specific genetic signature—the presence of a ‘good’ or ‘bad’ sequence—the macrolide can stop its production on the eukaryotic ribosome or not. This showed us, conceptually, that antibiotics can be used to selectively inhibit protein synthesis in human cells and used to treat human disorders caused by ‘bad’ proteins,” said Mankin.
The UIC researchers’ efforts serve as a testing ground for future studies.
“Now that we know the concepts work, we can look for antibiotics that are capable of binding in the unmodified eukaryotic ribosomes and optimize them to inhibit only those proteins that are bad for a human,” concluded Mankin.
Source:
Journal reference:
Svetlov, M. S, et al. (2021) Context-specific action of macrolide antibiotics on the eukaryotic ribosome. Nature Communications. doi.org/10.1038/s41467-021-23068-1.