Plastic was previously considered a manufacturing miracle as it was inexpensive to produce. However, it is hard to degrade and hence considered an environmental plague today, choking waterways and clogging landfills.
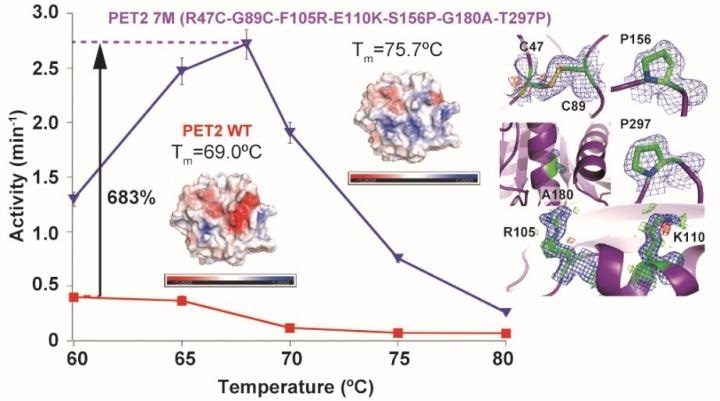
Mutations of a PET hydrolyzing enzyme PET2 resulted in a 6.7 °C increase in thermal stability and a 6.8-fold increase in PET hydrolytic activity. Tm represents melting (denaturation) temperature, and colors on enzyme structures show their surface charges (blue: positive, red: negative). Image Credit: NINS/IMS.
Japanese researchers sought help from nature to devise an approach to degrade plastic. The researchers engineered a protein that could bind to plastic particles and efficiently break them down, similar to the binding of a protein to cellulose in plants or to chitin in crustaceans to initiate decomposition.
The research findings were published on June 29th, 2021, in ACS Catalysis, a journal of the American Chemical Society.
Polyethylene terephthalate (PET) is produced and used in large quantities in modern society due to its low cost and ease of processing. However, in recent years, from the perspective of realizing a sustainable society, the complete recycling of PET in industry and the removal of PET from the natural environment have become global issues. To resolve these issues, it is very important to understand how to degrade PET efficiently.”
Ryota Iino, Study Author and Professor, Institute for Molecular Science, National Institutes of Natural Sciences
The scientists examined and engineered an enzyme cloned from a library of genetic materials collected from nature. Named PET2, the enzyme facilitated the degradation of PET by accelerating the reaction between chemical components in PET and water.
The researchers used single-molecule imaging analysis and disclosed that how the enzyme binds to the surface of PET limited the rate of degradation.
We also revealed that by introducing positive charges on the surface of PET-degrading enzyme, the binding rate to the PET surface can be increased.”
Ryota Iino, Study Author and Professor, Institute for Molecular Science, National Institutes of Natural Sciences
The positive charges favorably react on the PET surface, enhancing enzyme binding leading to effective degradation of the PET.
The scientists also discovered that engineered PET2 exhibited high thermal stability and highest activity at 68 °C, which is slightly lower than most residential kitchen ovens. However, at higher temperatures, it may be more effective as the PET’s molecular bonds become more flexible and breakable.
Our ultimate goal is to create a bacterium that can sense PET in the environment, move toward it, and degrade it.”
Ryota Iino, Study Author and Professor, Institute for Molecular Science, National Institutes of Natural Sciences
The created bacterium will then be able to transform the degraded PET into energy useful for other organisms, serving effectively as an automated recycling center for plastic. “In nature, chitin and cellulose are recycled in this way,” added Iino.
Source:
Journal reference:
Nakamura, A., et al. (2021) Positive Charge Introduction on the Surface of Thermostabilized PET Hydrolase Facilitates PET Binding and Degradation. ACS Catalysis. doi.org/10.1021/acscatal.1c01204.