Scientists have created an innovative artificial intelligence algorithm known as “scArches” for clinical applications. The algorithm performs an efficient comparison of patients’ cells with a reference atlas including cells of healthy individuals.
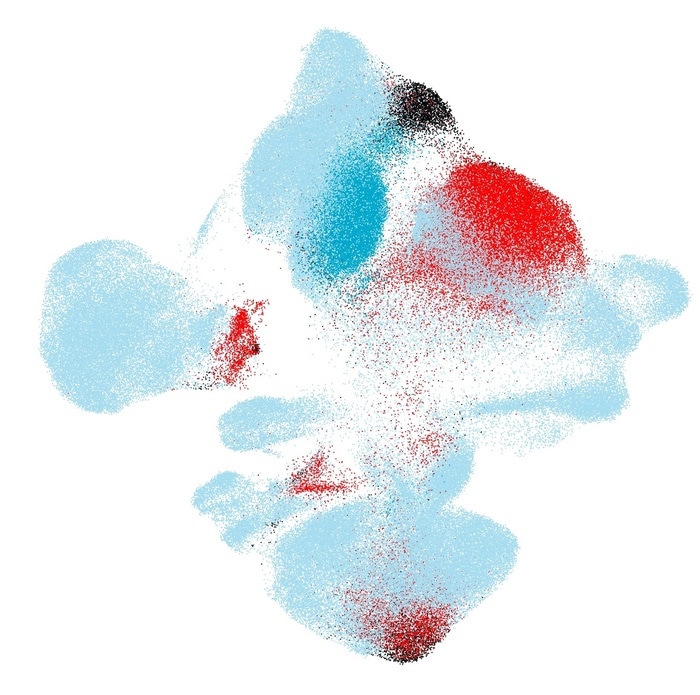
Mapping new cohorts of cells of healthy individuals and COVID-19 patients onto a healthy cells reference atlas (Light blue: Healthy reference patients. Blue: New healthy patients. Black: New moderate COVID-19 patients. Red: New severe COVID-19 patients.) Image Credit: Helmholtz Zentrum München.
Physicians can use this algorithm to identify cells in disease and prioritize them for personalized treatment of each patient.
The Human Cell Atlas—the largest, expanding single-cell reference atlas in the world—consists of references of millions of cells over organs, tissues, and developmental stages. Using these references, physicians can gain better insights into the impacts of environment, aging, and disease on a cell—and eventually perform better diagnosis and treatment for patients.
However, reference atlases pose their own challenges. Single-cell datasets may include measurement errors (batch effect), the worldwide availability of computational resources is limited, and raw data sharing is generally restricted legally.
Scientists from Helmholtz Zentrum München and the Technical University of Munich (TUM) created a new algorithm known as “scArches,” which stands for single-cell architecture surgery. Mohammad Lotfollahi, the leading scientist behind the algorithm, explained its biggest advantage as follows:
Instead of sharing raw data between clinics or research centers, the algorithm uses transfer learning to compare new datasets from single-cell genomics with existing references and thus preserves privacy and anonymity. This also makes annotating and interpreting of new data sets very easy and democratizes the usage of single-cell reference atlases dramatically.”
Mohammad Lotfollahi, Lead Scientist, Helmholtz Zentrum München
Example COVID-19
The scArches were applied by the team to investigate COVID-19 in many lung bronchial samples. Through single-cell transcriptomics, the cells of COVID-19 patients were compared with those of healthy references.
The algorithm could differentiate diseased cells from the references and thus allow users to identify the cells that need treatment, for both mild and severe COVID-19 cases. Biological differences between patients did not have any effect on the quality of the mapping process.
Our vision is that in the future we will use cell references as easily as we nowadays do for genome references. In other word, if you want to bake a cake, you usually do not want to try coming up with your own recipe—instead you just look one up in a cookbook. With scArches, we formalize and simplify this lookup process.”
Fabian Theis, Helmholtz Zentrum München
Source:
Journal reference:
Lotfollahi, M., et al. (2021) Mapping single-cell data to reference atlases by transfer learning. Nature Biotechnology. doi.org/10.1038/s41587-021-01001-7.