The fight against the COVID-19 pandemic is a worldwide scientific issue including the development of measures to combat the virus and its harmful effects. Apart from vaccines and drugs licensed by the health authorities, new therapies are needed to combat the viral infection once it occurs.
This new scientific advance is based on the use of in silico virtual screening, a computational analytical technique for selecting molecules that are candidates to future drugs in a chemical library. Image Credit: University of Barcelona.
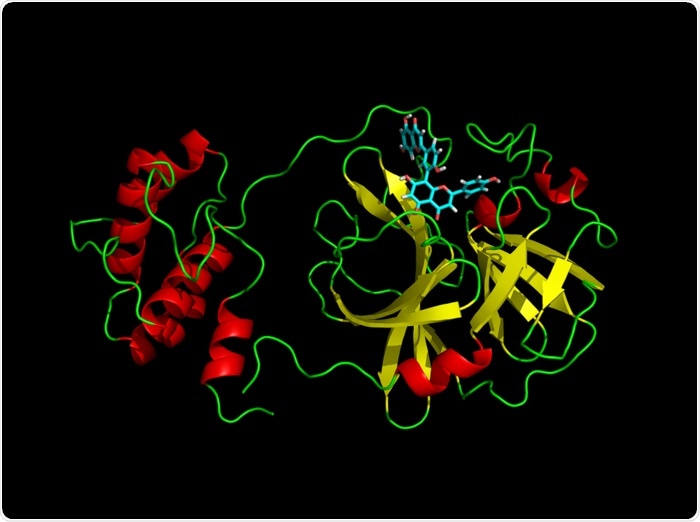
A variety of natural compounds capable of inhibiting Mpro, the main protease of SARS-CoV-2, was discovered in research published in the Journal of Chemical Information and Modeling. Mpro is a non-structural protein that plays an important role in the virus’s reproduction and transcription, and it is being looked at as a possible therapeutic target since its suppression might stop the virus from spreading.
Professor Jaime Rubio Martnez of the University of Barcelona’s Faculty of Chemistry and the Institute of Theoretical and Computational Chemistry (IQTC) conducted the research, which might help with the development of novel COVID-19 therapy techniques.
This new scientific breakthrough is based on the use of in silico virtual screening, a computer analytical approach for identifying and selecting chemicals that are candidates for future pharmacological relevance from databases.
Virtual screening: searching for future drugs
COVID-19’s therapeutic strategy now focuses on symptomatic therapy with anti-inflammatory drugs (such as dexamethasone or cytokine inhibitors) and antibiotics to treat secondary infections. The antiviral ritonavir was just licensed, but additional drugs are still needed.
The development of SARS-CoV-2-specific antiviral therapies is critical for global health, which has led to the employment of many techniques to find bioactive molecules that can be employed as therapeutic agents (including the available drugs and natural products). In this context, virtual screening is a dependable, quick, and effective method for identifying bioactive chemicals in massive chemical collections that are targeted against a specific molecular target.
The researchers used a molecular dynamics analysis to simulate a virtual screening of the natural product database Selleck—a chemical library with over 2,000 compounds—for a series of conformations of the SARS-CoV-2 Mpro.
As a consequence, the work specifies a series of typical structures and offers the characterization of the dynamic profile of Mpro protease in its apo form using conventional simulations (cMD) and Gaussian accelerated molecular dynamics (GaMD) simulations.
These structures were then utilized to perform ensemble docking, a bioinformatic approach for predicting and computing the most favorable site of contact between a ligand and its target protein using computer tools. Then, researchers used an iterative technique to lengthen the cMD of protein-ligand complexes and compute the free binding energy to find the most potential options.
Scientists chose eleven compounds based on the data and evaluated them in vitro for their potential to inhibit Mpro protease. Using the natural products database, five SARS-CoV-2 antiviral candidates were discovered as Mpro protease inhibitors.
Source:
Journal reference:
Rubio-Martínez, J., et al. (2022) Discovery of Diverse Natural Products as Inhibitors of SARS-CoV-2 Mpro Protease through Virtual Screening. Journal of Chemical Information and Modeling. doi.org/10.1021/acs.jcim.1c00951.