Why do genetically similar cells react to the same extrinsic stimuli, such as antibiotics, in such diverse ways? Mathematicians from KAIST and IBS have solved a long-standing puzzle by developing a new framework for understanding cell responses to various stimuli.
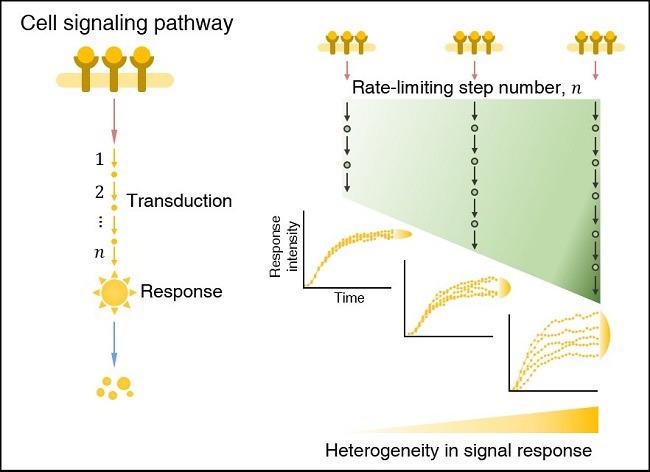 In a cell signaling pathway, the heterogeneity (i.e., cell-to-cell variability) in the signal response increases as the number of rate-limiting step number increases. Image Credit: Korea Advanced Institute of Science and Technology.
In a cell signaling pathway, the heterogeneity (i.e., cell-to-cell variability) in the signal response increases as the number of rate-limiting step number increases. Image Credit: Korea Advanced Institute of Science and Technology.
The scientists determined that when the effective length of the cell signaling route (i.e., the number of rate-limiting stages) grows, cell-to-cell variability in antibiotic stress response rises. This discovery might lead to the development of more effective chemotherapies to combat cancer cell fractionation induced by cell-to-cell variability.
Signal transduction mechanisms in human cells respond to a variety of external stimuli, including antibiotics and variations in osmotic pressure. Various biological responses occur in sequential order when an external input is recognized. As a result, necessary genes are expressed, enabling the cells to adjust to the disrupted external environment. Additionally, signal transduction results in a drug reaction (e.g., antibiotic resistance genes are expressed when antibiotic drugs are given).
Individual cell responses are extremely variable, even when identical external stimuli are recognized. As a result, persister cells form, which are very resistant to drugs. Many research has been performed to investigate the probable origins of this cell-to-cell variability. However, with present experimental approaches, the majority of intermediary signal transduction processes are unobservable.
Professor Jae Kyoung Kim of the KAIST Department of Mathematical Sciences and the IBS Biomedical Mathematics Group directed a team of researchers, including Dae Wook Kim and Hyukpyo Hong, to unravel the mystery using queueing theory and Bayesian inference methods. Scientists hypothesized a signal transduction mechanism in cells that are described by a queueing process.
Researchers used to construct Bayesian inference computational tools based on this MBI (the Moment-based Bayesian Inference method). This allows the signal transduction system to be studied without having to look at the intermediary steps directly. This research was published in the journal Science Advances.
The study team discovered that the number of rate-limiting stages in the signaling pathway grows as the number of cell-to-cell variability increases by evaluating experimental results from Escherichia coli using MBI.
The rate-limiting stages are the slowest (i.e., bottlenecks) in the sequential biochemical reaction tasks that make up cell signaling pathways, and they take up the majority of signaling time. Even in a population of genetically identical cells, the strength of the transduced signal becomes substantially diverse as the number of rate-limiting stages grows.
This discovery is likely to open up new avenues for research into cell heterogeneity in terms of antibiotic resistance, which is a major hurdle in cancer treatment.
“As a mathematician, I am excited to help advance the understanding of cell-to-cell variability in response to external stimuli. I hope this finding facilitates the development of more effective chemotherapies,” Professor Kim concluded.
Source:
Journal reference:
Kim, D. W., et al. (2022) Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: The rate-limiting step number. Science Advances.