Reviewed by Danielle Ellis, B.Sc.Jun 2 2022
Infectious-bacteria-killing molecular machines have been persuaded to reconsider their goal.
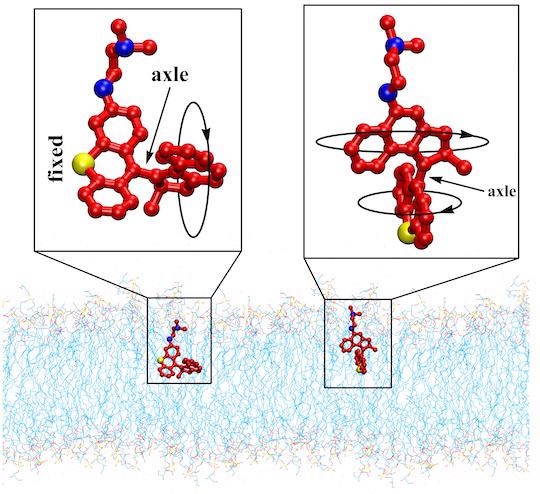 The schematics show two variants of light-activated molecular machines developed at Rice University that drill into and destroy antibiotic-resistant bacteria. The machines could be useful to fight infectious skin diseases. Image Credit: Tour Research Group/Rice University
The schematics show two variants of light-activated molecular machines developed at Rice University that drill into and destroy antibiotic-resistant bacteria. The machines could be useful to fight infectious skin diseases. Image Credit: Tour Research Group/Rice University
The current edition of Rice University’s nanoscale drills is operated by visible light rather than ultraviolet (UV), as in previous generations. These have also been shown to be effective at killing bacteria in real-world diseases.
Rice chemist James Tour and his team successfully demonstrated six molecular machine variations. In as little as two minutes, they all punched holes in the membranes of gram-negative and gram-positive bacteria.
For microorganisms with no natural defenses against mechanical intruders, resistance proved fruitless. That means they are unlikely to develop resistance, which means they could be used to combat bacteria that have developed resistance to standard antibiotic treatments over time.
I tell students that when they are my age, antibiotic-resistant bacteria are going to make COVID look like a walk in the park. Antibiotics will not be able to keep 10 million people a year from dying of bacterial infections. But this really stops them.”
James Tour, Chemist, Rice University
The groundbreaking research, conducted by Tour and Rice graduate Ana Santos, was published in Science Advances.
The Rice lab has been perfecting its molecules for years because prolonged exposure to UV can be harmful to people. The new edition obtains its energy from 405 nanometer light, which spins the molecules’ rotors at a rate of 2 to 3 million times per second.
Several researchers have suggested that light at that wavelength has weak antibacterial capabilities on its own, but adding molecular machines boosts it, according to Tour, who believes that bacterial diseases like those encountered by burn victims and persons with gangrene will be the first targets.
The machines are built on Nobel Prize-winning Bernard Feringa’s work in 1999 when he created the first molecule with a rotor and got it to spin reliably in one direction. In a 2017 Nature journal, Tour and his colleagues described their advanced drills.
The novel compounds’ capacity to kill bacteria swiftly was confirmed in the Rice lab’s first experiments on burn wound infection models, particularly methicillin-resistant Staphylococcus aureus, a frequent cause of skin and soft tissue infections that killed over 100,000 people in 2019.
By incorporating a nitrogen group, the researchers were able to accomplish visible light activation.
“The molecules were further modified with different amines in either the stator (stationary) or the rotor portion of the molecule to promote the association between the protonated amines of the machines and the negatively charged bacterial membrane,” said Liu, now a researcher at Arcus Biosciences in California.
The robots also efficiently break up biofilms and persister cells, which go dormant to evade antibacterial treatments, according to the researchers.
Even if an antibiotic kills most of a colony, there are often a few persister cells that for some reason don’t die. But that doesn’t matter to the drills.”
James Tour, Chemist, Rice University
The new devices, like previous iterations, claim to resurrect antibacterial medications that have been deemed ineffective.
“Drilling through the microorganisms’ membranes allows otherwise ineffective drugs to enter cells and overcome the bug’s intrinsic or acquired resistance to antibiotics,” said Santos, who is in her third year of a postdoctoral global fellowship that introduced her to Rice for two years and is now progressing at the Health Research Institute of the Balearic Islands in Palma, Spain.
By attaching bacterium-specific peptide tags to the drills and directing them toward pathogens of interest, the lab hopes to improve bacteria targeting and reduce damage to mammalian cells.
“But even without that, the peptide can be applied to a site of bacterial concentration, like in a burn wound area,” Santos concluded.
Source:
Journal reference:
Santos, A. L., et al. (2022) Light-activated molecular machines are fast-acting broad-spectrum antibacterials that target the membrane. Science Advances. doi.org/10.1126/sciadv.abm2055.