Reviewed by Danielle Ellis, B.Sc.Aug 9 2022
Researchers at Johns Hopkins Medicine claim to have discovered significant molecular variations between cancer cells that adhere to an initial tumor and those that spread out to develop distant tumors while researching the fatal form of breast cancer known as triple negative.
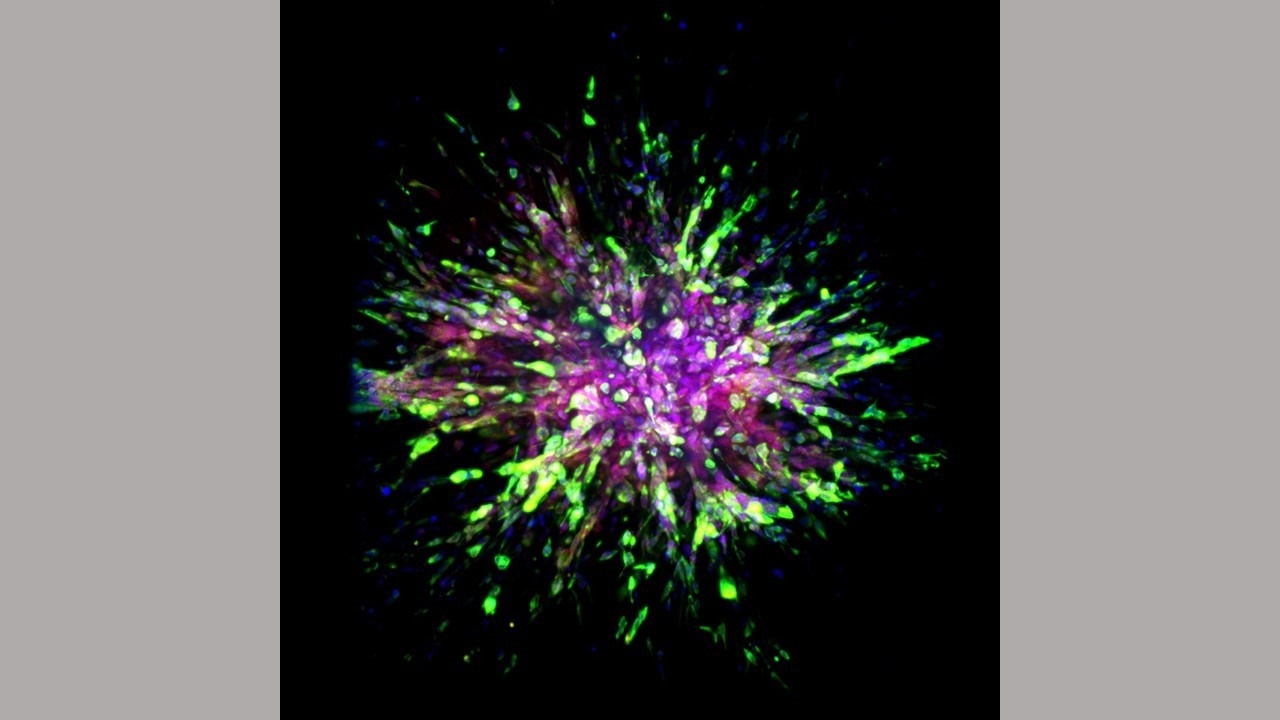 A triple-negative breast cancer organoid invades collagen tissue. Image Credit: Eloïse Grasset, Johns Hopkins Medicine.
A triple-negative breast cancer organoid invades collagen tissue. Image Credit: Eloïse Grasset, Johns Hopkins Medicine.
The study, which included mouse models and human tissues, may pave the way for the creation of fresh therapies that focus on these genetic variances.
On August 3rd, 2022, the study was published in the journal Science Translational Medicine.
We have long needed new treatment targets and options for triple negative breast cancers. These cancers often return within three years of diagnosis, and treatments used for other breast cancers don’t typically work for triple negative.”
Andrew Ewald PhD, Director, Department of Cell Biology, Johns Hopkins University School of Medicine
Andrew Ewald is also the Virginia DeAcetis Professor in Basic Cancer Research and co-leader of the Cancer Invasion and Metastasis Program at the Johns Hopkins Kimmel Cancer Center.
Triple negative breast cancer accounts for an estimated 10–20% of the 280,000 cases of breast cancer that are diagnosed annually in the United States. The rate is higher among African American women, who are twice as likely as other women to develop this type of cancer.
This form of cancer is particularly deadly because it lacks molecular signals on its surface that communicate with the hormones progesterone and estrogen as well as the cancer-promoting protein Her2-neu. The majority of breast cancer treatments available today target those markers, making them ineffective for those with triple-negative tumors.
The research team used three different types of cells for the current study: mouse models, human cancers implanted into mice, and samples of both primary and metastatic tissues taken from eight patients treated at The Johns Hopkins Hospital. They examined molecular variations between the initial, or primary, triple-negative breast cancer sites, and areas where it spread or metastatic sites.
To distinguish distinctions between the genetic expression patterns of primary and metastatic cancers, the researchers combined machine learning, cellular imaging, and biochemical analysis.
The bad news from our study is that cells from metastatic sites are super optimized for migration and resisting treatment. The good news is that we identified several proteins called transcription factors that these cells require to handle the challenges of migrating and thriving at metastatic sites, and we may be able to design new therapies that target these transcription factors.”
Andrew Ewald PhD, Director, Department of Cell Biology, Johns Hopkins University School of Medicine
More particularly, Ewald, Johns Hopkins postdoctoral fellow Eloïse Grasset PhD, and other members of the research team discovered several distinctive characteristics in the cells of mice whose breast cancers were engineered to be triple negative and mice implanted with human triple negative breast cancer tumors.
The researchers discovered that triple negative breast cancer cells acquire two biological properties: greater mobility and survival—when they infect other tissues on their way to another section of the body.
To achieve this, breast cancer cells acquire the vimentin cellular skeleton protein, which improves the capacity of so-called mesenchymal cells to migrate and generate new cells. Mesenchymal cells are a type of cell that are generally found in bones and bone marrow.
A protein called E-cadherin, which is generally present in epithelial cells that line the ducts and coverings of organs and constantly renew themselves, is produced by triple negative breast cancer cells to help them survive.
Scientists categorize the biological state of triple negative breast cancer cells as so-called hybrid epithelial mesenchymal (EMT) cells when they exhibit these survival and migratory characteristics.
The researchers worked with Elana Fertig PhD, division director and associate director of quantitative sciences and co-director of the Convergence Institute at the Johns Hopkins Kimmel Cancer Center, to follow the molecular patterns of individual cells in cell assays that simulate invasion out of the primary tumor and formation of a colony in a metastatic site. This allowed them to take a closer look at molecules involved in hybrid EMT states.
The computational team led by Fertig used machine learning methods to identify trends in each cell’s expression of RNA, a DNA cousin involved in protein synthesis. The majority of metastatic cells, the researchers discovered, transform into the hybrid EMT state, which is more migratory and more resilient.
Ewald’s group examined both primary tumors and tissues from the same patients’ metastatic sites to validate similar states in samples from eight patients with triple negative malignancies.
Five transcription factors (Grhl2, Foxc2, Zeb1, Zeb2, and Ovol1), which encourage the synthesis of proteins involved in either colony formation or cancer cell invasion, were expressed by the majority of metastatic cells.
The molecular differences between metastatic and primary tumors are likely the reason why metastatic tumor cells are so resistant to current treatments.”
Andrew Ewald PhD, Director, Department of Cell Biology, Johns Hopkins University School of Medicine
The team is investigating whether the same molecular and cellular alterations occur in other malignancies, such as those of the colon, adrenal glands, stomach, and small intestine, and how to inhibit the genes or proteins produced by the transcription factors to stop the spread of metastatic disease.
Source:
Journal reference:
Grasset, E. M., et al. (2022) Triple-negative breast cancer metastasis involves complex epithelial-mesenchymal transition dynamics and requires vimentin. Science Translational Medicine. doi.org/10.1126/scitranslmed.abn7571.