The most essential foundation of life on Earth is photosynthesis. Plants and single-celled algae use sunlight’s energy to turn it into sugar and biomass. This mechanism results in the release of oxygen. Plant biotechnologists and structural biologists from Münster and Stockholm (Sweden) have solved the structure of a novel protein complex that catalyzes energy conversion pathways in photosynthesis.
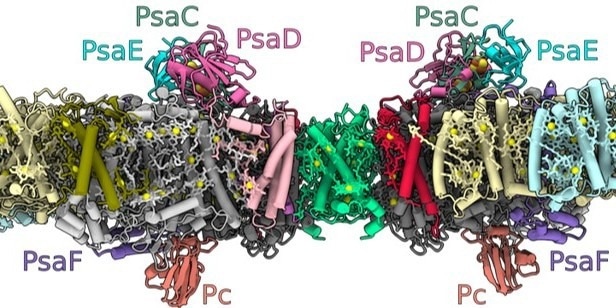 The structure of the photosystem I dimer embedded in the thylakoid membranes within the chloroplasts, where the light-driven photosynthesis process takes place (inside of the membrane: lumen, outside: stroma). The letter combinations denote different proteins that the researchers have identified. Image Credit: © Nature Plants
The structure of the photosystem I dimer embedded in the thylakoid membranes within the chloroplasts, where the light-driven photosynthesis process takes place (inside of the membrane: lumen, outside: stroma). The letter combinations denote different proteins that the researchers have identified. Image Credit: © Nature Plants
The photosystem I protein complex is recognized as a single protein complex (monomer) in plants. Prof. Michael Hippler of the University of Münster and Prof. Alexey Amunts of the University of Stockholm led a group of scientists who demonstrated for the first time that two photosystem I monomers in plants can combine as a dimer, and they described the molecular structure of this new type of molecular machine.
The findings, which have been reported in the journal “Nature Plants,” offer molecular insights into the process of photosynthesis with a level of precision that has never before been attained. They might make it possible to utilize photosystem I’s reductive force—or readiness to give up electrons—more effectively in the future. For instance, they might make it possible to generate hydrogen as an energy source.
The background
Photosystems I and II are two photosynthesis complexes that function most effectively when exposed to light of various wavelengths. Light energy is absorbed by photosystems I and II, where it drives the movement of electrons inside the molecular “photosynthetic machine” and the transformation of light energy into chemical energy.
Electrons from photosystem I are transferred to the protein ferredoxin during the process. Ferredoxin in green algae can transfer electrons generated during photosynthesis to an enzyme called hydrogenase, which then creates molecular hydrogen. This molecular hydrogen is thus created by the input of light energy, implying that it is renewable and may be useful as a future source of energy.
The investigators wondered, “How does the production of photosynthetic hydrogen relate to the structural dynamics of the monomer and dimer photosystem I?”
The results in detail
The green alga Chlamydomonas reinhardtii’s photosystem I homodimer is made up of 40 protein subunits and 118 transmembrane helices, which together provide a structure for 568 photosynthesis pigments.
With the help of cryogenic electron microscopy, the researchers demonstrated that the absence of PsaH and Lhca2 subunits result in a head-to-head orientation of monomer photosystem I (PSI) and its linked light-harvesting proteins (LHCI). This dimerization is made possible by the light-harvesting protein Lhca9.
In the research, the scientists characterize the most accurate PSI-LHCI model to a resolution of 2.3 Ångström (1 Ångström corresponds to one ten-millionth of a millimeter); they also assign the accurate identity and orientation to all pigments, in addition to 621 water molecules that have an impact on the pathways for energy transmission.
The genetically driven down-regulation of the component Lhca2 causes the double mutant to produce hydrogen very effectively in conjunction with the deletion of a second gene (pgr5).
The depletion of Lhca2 promotes the formation of PSI dimer, and so we suggest that the hydrogenase may favor the targeting of photosynthetic electrons from the PSI dimer, as we proposed in our earlier work. The structure of the PSI dimer enables us to make targeted genetic modifications in order to test the hypothesis of improved hydrogen production through the PSI dimer.”
Michael Hippler, Professor, University of Münster
Source:
Journal reference:
Naschberger A., et al. (2022) Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex. Nature Plants. doi.org/10.1038/s41477-022-01253-4.