The mTOR protein is essential for cell growth, proliferation, and survival. Its activity fluctuates according to nutrient availability and growth factors like hormones. This protein has been linked to several illnesses, including cancer, where its activity is typically increased.
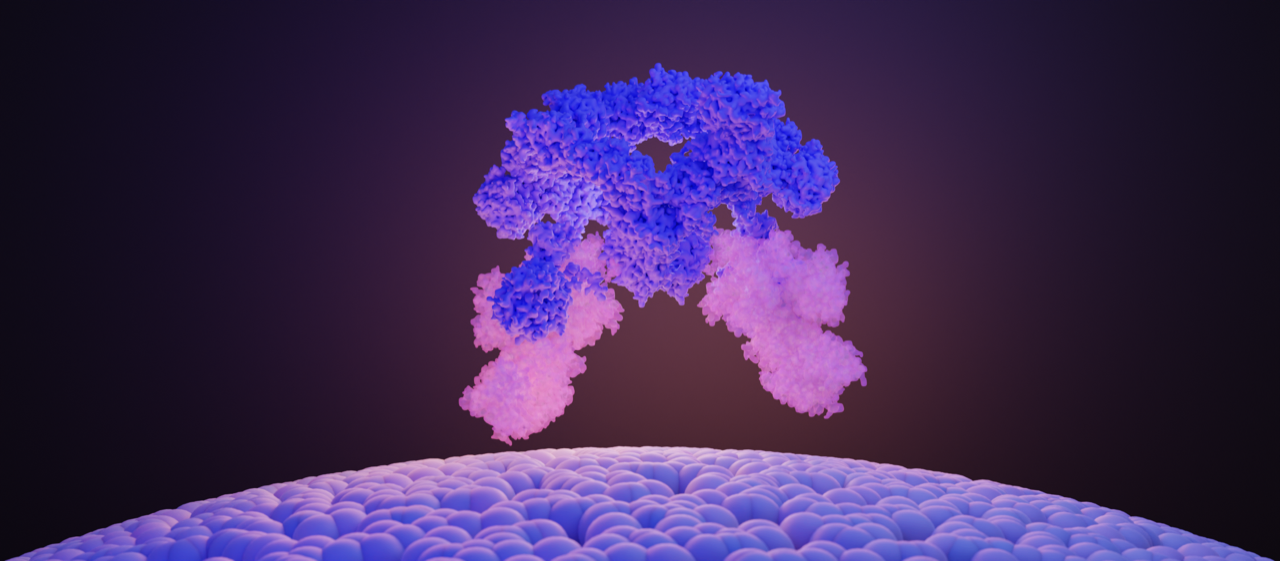 The SEA complex is composed of a cage-like core (SEACAT, blue) that regulates the activity of the wings (SEACIT, white and bright). Image Credit:© Ciencia Graficada.
The SEA complex is composed of a cage-like core (SEACAT, blue) that regulates the activity of the wings (SEACIT, white and bright). Image Credit:© Ciencia Graficada.
Researchers from the University of Geneva (UNIGE), Martin Luther University (MLU) in Halle-Wittenberg in Germany, and the recently opened Dubochet Center for Imaging (UNIGE-UNIL-EPFL) have worked together to identify the structure of the SEA complex, an interdependent group of proteins, which controls mTOR to better comprehend how it is regulated.
The finding of this structure improves the knowledge of how cells perceive nutrient levels to control their growth. The study is published in the journal Nature.
The mTOR protein (mammalian target of rapamycin) is the master regulator of cell proliferation from yeast to humans.
This protein responds to environmental cues like nutrients and hormones and controls several critical cellular functions like protein and lipid synthesis, energy production by mitochondria, and cell structure organization. mTOR activity disruptions are the root cause of many disorders, including diabetes, epilepsy, obesity, and several types of cancer.
Two opposing functions in the same complex
Robbie Loewith’s laboratory, Professor in the Department of Molecular and Cellular Biology at the UNIGE Faculty of Science and Director of the National Center for Competence in Research in Chemical Biology, is focused on the regulation of mTOR, particularly the SEA complex, which is a direct sensor of nutrients and regulate mTOR activity.
The SEA complex is made up of eight different proteins. One component of the SEA complex (SEACIT) is engaged in mTOR inhibition, while the other component (SEACAT) is involved in mTOR activation.
The SEACIT subcomplex inhibits the mTOR protein, which prevents cell development in the absence of nutrients. The SEACIT subcomplex, which can no longer inhibit the mTOR protein, is considered to be blocked by the SEACAT subcomplex in the presence of nutrients.
The central controller can then use other mechanisms, such as inducing the creation of proteins and lipids, to activate cell growth. Uncertainty still exists around how SEACAT controls SEACIT.
Determining structure to understand function
The investigators set out to identify the structure of the SEA complex to figure out the interactions between the proteins and to further comprehend how they work.
The SEA complex was biochemically separated from the rest of the cell’s components, and then the researchers employed cryo-electron microscopy (cryo-EM) and the technologies of the Dubochet Center for Imaging of UNIGE, UNIL, and EPFL to determine its molecular structure.
By freezing the samples very quickly at −180 °C, cryo-EM allows us to obtain the structure of the proteins in their original state, i.e., in their functional three-dimensional form.”
Lucas Tafur, Study First Author and Researcher, Department of Molecular and Cellular Biology, University of Geneva
SEACAT is necessary but not sufficient
The complex’s various biochemical functions were subsequently examined in a laboratory setting. Even though the SEACAT subcomplex remains active (as it is when nutrients are present), the researchers discovered that the SEACIT subcomplex is still active and capable of inhibiting mTOR.
This result is very unexpected since SEACAT has long been described as the direct inhibitor of SEACIT. We, therefore, expected SEACIT to be inactive in the presence of active SEACAT. Our results show that SEACAT acts more as a scaffold for the recruitment of other regulatory proteins and that its presence is therefore necessary but not sufficient for the inhibition of SEACIT.”
Robbie Loewith, Study Last Author, University of Geneva
The discovery of the structure of the SEA complex has enabled the identification of missing links in the mTOR regulatory cascade.
Of course, we now need to identify the as yet unknown partners that associate with this complex. These new factors could prove to be therapeutic targets for tumors where mTOR activity is exacerbated.”
Lucas Tafur, Study First Author and Researcher, Department of Molecular and Cellular Biology, University of Geneva
Source:
Journal reference:
Tafur, L., et al. (2022) Cryo-EM structure of the SEA complex. Nature. doi.org/10.1038/s41586-022-05370-0.