The best method now available for researchers to isolate a specific microorganism with a given metabolic function from an environment is to collect a sample of cells, culture them, and then test them for the desired cell functions.
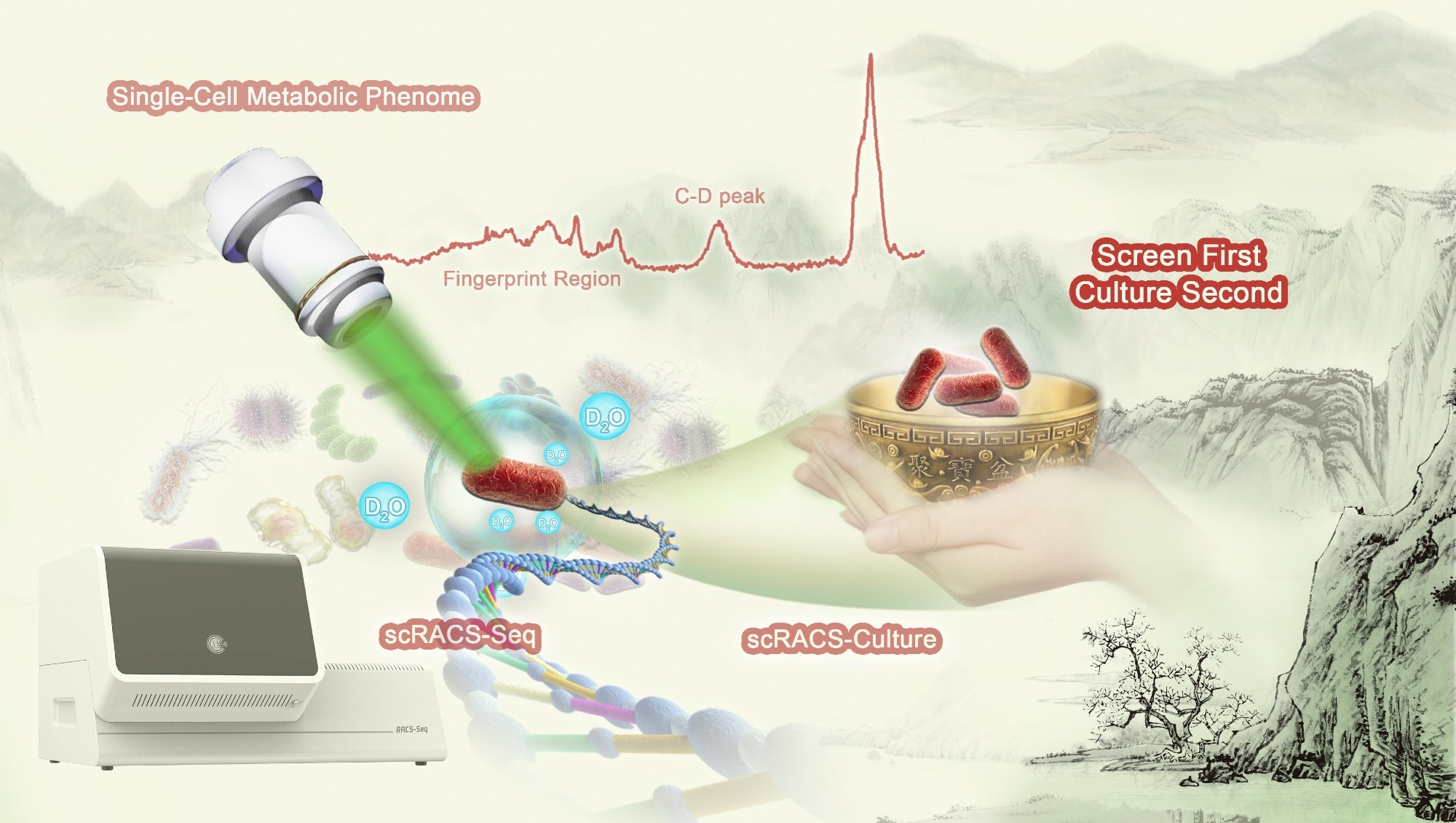 A complete workflow of scRACS-Culture for bioresource mining from environmental microbiomes. Image Credit: Yang Liu.
A complete workflow of scRACS-Culture for bioresource mining from environmental microbiomes. Image Credit: Yang Liu.
This approach has various drawbacks. First, most cells in nature have not been discovered by scientists, restricting the results to earlier grown cells. Second, how cells act in a test tube does not always correspond to how they behave in nature or in situ. This can make identifying the correct cell with the correct metabolic functions difficult.
Scientists from the Chinese Academy of Sciences’ Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) have suggested a new method called single-cell Raman-activated sorting and cultivation, or scRACS-Culture, to identify and harvest cells from the environment by screening the cells initially and then culturing them.
The research was published on October 30th, 2022, in the journal ISME Communications.
Function-based mining of microbes from nature has traditionally employed a ‘culture first, screen second’ strategy, which has several limitations. By employing phosphate-solubilizing microbes as a model, we introduced a ‘screen first’ strategy called single-cell Raman-activated sorting and cultivation.”
Xiaoyan Jing, Study First Author and Senior Engineer, Single-Cell Center, Qingdao Institute of Bioenergy and Bioprocess Technology
The researchers established the viability of scRACS-Culture by mining microorganisms for in situ organic-phosphate solubilizing activity from an urban wastewater treatment plant using a unique instrument called RACS-Seq.
This is a real-world application for this technology since organic-phosphate solubilizing strains are in great demand because they can reduce pollution in water bodies and soil, boost crop nutrient absorption, enhance fertilizer efficacy, and decrease the use of chemical fertilizers.
The researchers employed the scRACS-Culture approach to directly mine such organisms from a wastewater sample by observing their metabolic rate when only organic-phosphate substrates were available. They highlighted that such a culture-independent, “screen-first” technique has the advantage of screening all cells in a microbiome rather than just those that can be cultivated.
Furthermore, this technique may assess the cell's in-situ function, which is frequently more important than the function of a pure culture in a test tube. It is also applicable to a large range of important cellular metabolic functions.
Because this technique can measure a wide range of metabolic phenotypes in a fluorescence-probe-free manner, it should greatly expand the use of function-driven single-cell technologies in microbiome science and industries.”
Yanhai Gong, Study Co-First Author and Assistant Research Fellow, Single-Cell Center, Qingdao Institute of Bioenergy and Bioprocess Technology
Researchers anticipate an approach that unlocks the target cell’s nutrient needs via metabolic reconstruction of its single-cell genome to boost the success rate of scRACS-Culture. The insights gained can subsequently be used to optimize the culture medium used to grow these yet-to-be-cultured single-cells into important live-bacteria cultures for animal and plant health, as well as environmental remediation.
According to Prof. Jian Xu of QIBEBT’s Single-Cell Center, who spearheaded the study, the group plans to increase the scale and throughput of the technology in the future to efficiently mine “probiotics” from a wide range of ecosystems.
Source:
Journal reference:
Jing, X., et al. (2022) Single-cell Raman-activated sorting and cultivation (scRACS-Culture) for assessing and mining in situ phosphate-solubilizing microbes from nature. ISME Communications. doi.org/10.1038/s43705-022-00188-3.