The structure of the influenza replication machinery and how it interacts with cellular proteins have been solved by a team of Oxford University scientists using a variety of techniques at Diamond Light Source. This study contributes to the understanding of influenza replication and the virus’s capacity for host-specific adaptation.
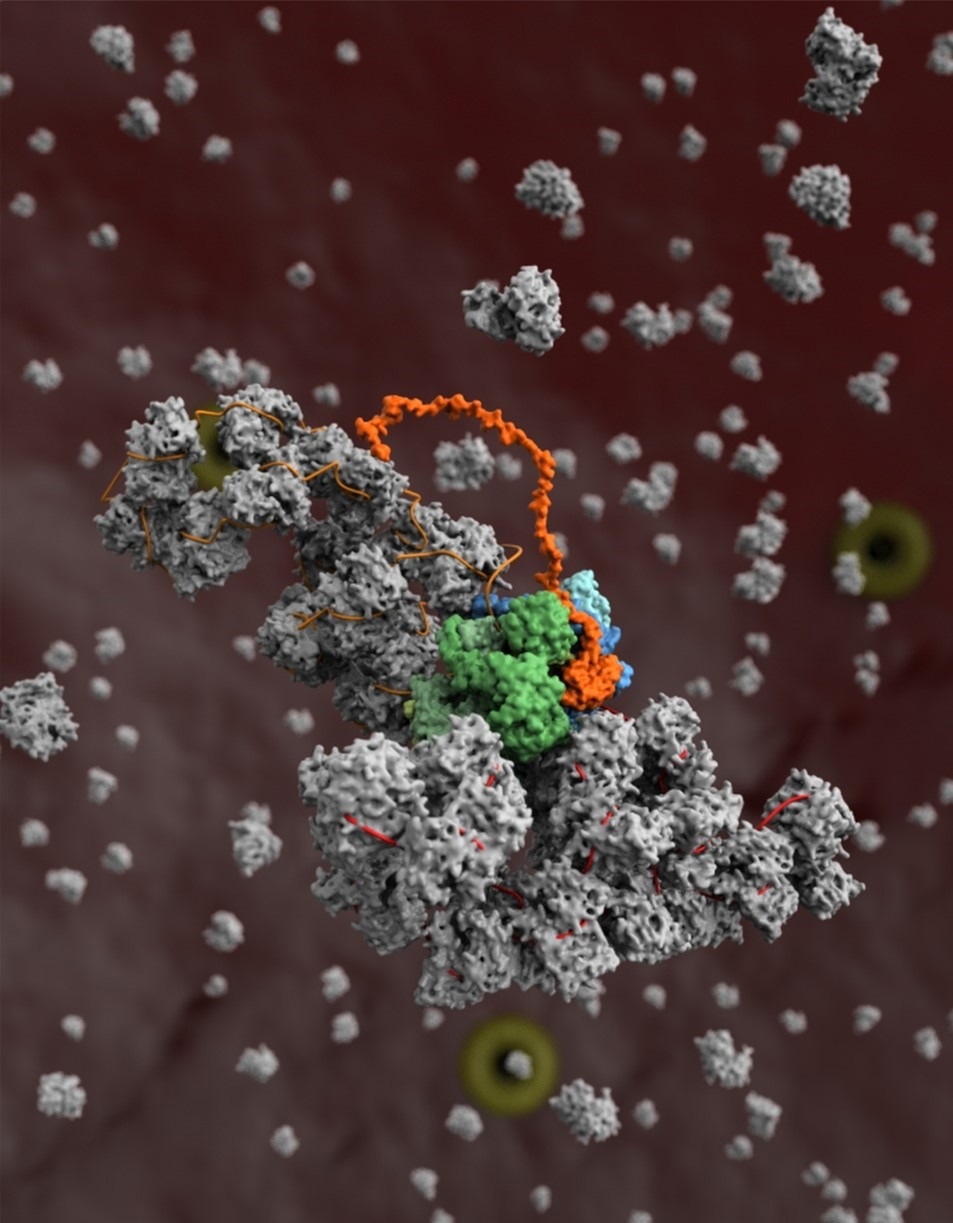 A model of the influenza virus replication machinery in the process of the viral RNA polymerase (blue) copying a viral RNA genome segment (red) in complex with a second copy of the polymerase (green) and host protein ANP32A (orange), and the assembly of the newly synthesized RNA segment (orange) into a ribonucleoprotein complex (grey). Image Credit: Diamond Light Source
A model of the influenza virus replication machinery in the process of the viral RNA polymerase (blue) copying a viral RNA genome segment (red) in complex with a second copy of the polymerase (green) and host protein ANP32A (orange), and the assembly of the newly synthesized RNA segment (orange) into a ribonucleoprotein complex (grey). Image Credit: Diamond Light Source
In addition to spreading seasonally, influenza can spread from animals to people and cause a pandemic. Researchers are piecing together how influenza sabotages human and animal cells for its replication by closely examining the virus’ replication cycle.
A recent study published in Trends in Microbiology describes the discoveries made possible by X-Ray crystallography, small angle X-Ray scattering (SAXS), cryo-EM, and Diamond’s electron Bio-Imaging Centre (eBIC).
New potential drug targets for the creation of innovative antiviral drugs to prevent influenza virus replication have been identified due to these structural insights.
The influenza virus replicates its genome by synthesizing its own RNA polymerase and storing its genes in RNA. In addition to replication, this viral polymerase performs a variety of other tasks that have been clarified by joint study across Diamond.
These investigations demonstrate that the polymerase controls the timing of transcription, the initial stage of protein synthesis, and replication, which can only start after viral proteins have been created. The research demonstrates how the polymerase interacts with the cellular protein ANP32A and makes use of it to conceal viral RNA from immune system detection.
The influenza A viruses that are currently in circulation are believed to be the evolutionary offspring of the virus that caused the 1918–1919 global pandemic, which resulted in 50–100 million fatalities globally.
Normally, influenza viruses can only infect a certain species of an animal host, such as birds, and must undergo certain changes to spread to other species, such as humans.
The 1918 influenza virus is regarded as the “founder virus” that provided viral genome fragments for all subsequent epidemic and pandemic strains, and it is likely to have spread from waterfowl to humans.
The team analyzed the structure of the 1918 pandemic influenza virus’ polymerase and located sites on its surface where the polymerase is vulnerable to inhibition in a study that was published earlier this year. This in turn can assist in locating and validating potential drug research targets.
This study is essential to comprehending how ANP32A contributes to the host-jumping barrier. Due to the significant differences in ANP32A between humans and birds, avian and animal influenza viruses have evolved to be less similar. Diamond’s structural biology research sheds light on the possibility of the pandemic spread of certain flu strains.
The researchers discovered that a change in the viral polymerase of avian influenza may allow it to interface with human ANP32A, enabling that strain of bird flu to infect human hosts by interacting with human ANP32A.
Large protein complexes can be difficult to structurally characterize, and the influenza replication complex was no exception. The structure of the viral polymerase was determined in nearly atomic detail by X-Ray crystallography at beamlines I03 and I24, which showed that the individual polymerases pair together to form dimers.
At beamline B21, a structural method known as SAXS was used to complement the crystal structure of the dimers and show how crucial dimer formation is to the polymerases’ ability to function.
According to the theory put forth by the researchers, single RNA polymerases initially engage in transcription during an infection before switching to replication only after forming dimers with other single polymerases.
Cryo-EM has allowed us to begin to look at very interesting protein complexes that we would find impossible to grow crystals of in the lab.”
Jonathan Grimes, Professor, Structural Virology, University of Oxford
Cryo-EM studies of the interactions between the viral polymerase and the RNA revealed that one polymerase in the dimer duplicates the viral genome while the other covers the newly generated RNA in viral proteins to protect it from immune sensors.
It is interesting to note that influenza subverts the cellular protein ANP32A to stabilize the dimers and help coat and conceal viral RNA to evade immune detection.
Grimes added, “Diamond democratizes science. The fact that all of these techniques exist in one place and are available to the scientific community is a hugely valuable resource. These world class cutting-edge facilities are freely available to scientists from universities and institutes across the UK and EU with interesting and important biological questions.”
These studies help us to identify and validate targets for drug discovery. We are hoping that the new insights generated into workings of the influenza virus transcription machinery using the technologies at Diamond will eventually lead to novel antivirals targeting the influenza polymerase.”
Ervin Fodor, Study Corresponding Author and Professor, University of Oxford
Source:
Journal reference:
Zhu, Z., et al. (2022). A structural understanding of influenza virus genome replication. Trends in Microbiology. doi.org/10.1016/j.tim.2022.09.015