A research team guided by Professor Xiang David Li of The University of Hong Kong’s (HKU) Department of Chemistry created a new chemical tool to unveil how bacteria adjust to the host environment and control host cells. This tool could be used to explore bacterial interactions with the host in real-time during an infection, which other methods cannot easily achieve.
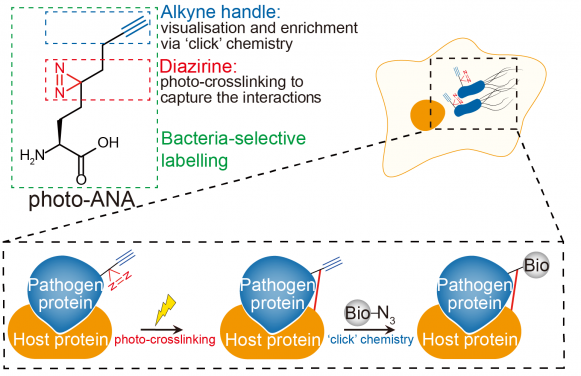 The figure shows the profiling of host-pathogen interactions. Photo-ANA is an unnatural amino acid containing an alkyne group and a diazirine group. After labeling bacterial during infection, photo-ANA enables the capture and purify of the targeted host proteins to unravel infection mechanisms. Image Credit: Image modified from an original illustration of Li et al., 2023 Nature Chemical Biology.
The figure shows the profiling of host-pathogen interactions. Photo-ANA is an unnatural amino acid containing an alkyne group and a diazirine group. After labeling bacterial during infection, photo-ANA enables the capture and purify of the targeted host proteins to unravel infection mechanisms. Image Credit: Image modified from an original illustration of Li et al., 2023 Nature Chemical Biology.
The observations were recently published in Nature Chemical Biology, a prestigious international scientific journal.
Despite having fewer than a hundred pathogenic bacteria, they pose a significant threat to human health around the world. Infection with Mycobacterium tuberculosis, for instance, causes tuberculosis, which kills over one million people each year. It was the world's deadliest infectious disease until COVID-19 surpassed it.
Despite effective antibiotic treatments, multidrug-resistant tuberculosis is becoming a global problem. As a result, a better understanding of how bacteria infect their human hosts is critical for developing new drugs and therapies.
When bacteria encounter their host (for example, human cells), they release “assassins” (virulence factor proteins) that “hijack” major protein players in the host to cause havoc during an invasion.
As a result, understanding bacterial infections requires investigating which virulence factors bacteria secrete and which host proteins are targeted. However, identifying these key players among the “crowded streets” can be extremely difficult (excessive host cellular matrix).
To address this issue, Professor Li’s group created photo-ANA, a multifunctional unnatural amino acid that only labels proteins of engineered bacteria but not the host during infection. Photo-ANA can conjugate with fluorescence or biotin via a Nobel Prize-winning chemical reaction (“click” chemistry), allowing for the visualization and enrichment of labeled bacterial proteins from the complex host environment.
As a result, Photo-ANA acts as an “undercover agent,” gathering intelligence and tagging all of the bacteria’s assassins. More importantly, photo-ANA contains a diazirine group that, when exposed to ultraviolet (UV) light, can “handcuff” the bacterial virulence proteins to their host target proteins, catching them in the act.
Professor Li’s group used photo-ANA to profile the adaptation of Salmonella, bacteria that can cause severe diarrhea, to the host environment, revealing the comprehensive interplay between Salmonella and the host during distinct infection stages, identifying known interactions as well as some newly discovered interactions. Furthermore, the photo-ANA-based approach is easily adaptable to other pathogenic bacteria and even fungi.
Researchers can now explore the activity of bacteria inside the host in real-time using this new chemical tool. In the future, it might be possible to use this tool to interpret the hidden interactions of deadly bacteria with the host as well as the mechanisms of multidrug-resistant superbugs, which will help better understand infectious diseases and inspire new treatments.
Source:
Journal reference:
Li, X.-M., et al. (2023) Photo-ANA enables profiling of host–bacteria protein interactions during infection. Nature Chemical Biology. doi.org/10.1038/s41589-022-01245-7.