A diverse community of several bacterial species that are crucial to human health constitute the gut microbiome. Changes in the gut microbiota have been linked to a range of diseases recently, most notably colorectal cancer (CRC), according to researchers across a variety of domains.
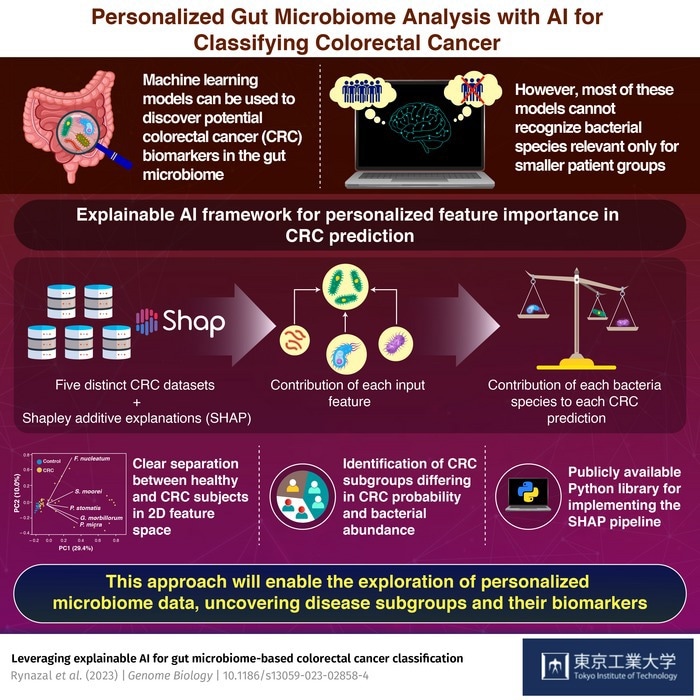
Image Credit: Tokyo Tech
Numerous studies have shown that the development of CRC is often correlated with a higher abundance of certain bacteria, such as Fusobacterium nucleatum and Parvimonas micra.
Based on these discoveries, scientists have created several artificial intelligence (AI) models to aid in the determination of which bacterial species are helpful as CRC biomarkers.
However, the majority of these models rely on what is referred to as “global explanations,” which means that they can only take all of the input data into account when making predictions. These models are therefore unable to locate bacterial species that could act as pertinent CRC biomarkers for smaller, less representative patient groups.
To solve this constraint, a research team from the Tokyo Institute of Technology (Tokyo Tech) in Japan decided to use a new strategy. The scientists used an explainable AI framework that offers local, as opposed to global, reasons for its CRC predictions, as described in their study, which was just published in Genome Biology.
Local explanation techniques make it possible to discover the most contributing bacteria for each individual CRC patient, enabling us to examine inter-individual differences between subjects within a disease group.”
Takuji Yamada, Study Main Author and Associate Professor, Tokyo Institute of Technology
The researchers employed a framework dubbed “Shapley additive explanations” (SHAP), which was inspired by an idea from a game theory known as the Shapley value. The Shapley value, in simple terms, explains how compensation should be divided among the participants of a coalition or group.
Similarly, the researchers employed SHAP in their investigation to determine the contribution of each bacterial species to each individual CRC prediction.
The researchers observed that projecting the SHAP values into a two-dimensional (2D) space allowed them to see a distinct demarcation between healthy and CRC subjects using this method and data from five CRC datasets. Clustering this 2D data produced four separate groupings of CRC subjects, each with a different CRC probability and associated bacteria.
Furthermore, the researchers discovered that people in the CRC subgroups with the highest CRC risk always had an enhanced population of bacteria linked with CRC. Most notably, the results were similar across the five datasets, demonstrating the method’s broad applicability.
With these encouraging findings, the team expects its method to make significant contributions to the gut microbiome research community.
Yamada further added, “Considering the increasing use of machine learning in microbiome–disease association studies, our novel method could be beneficial for a more personalized microbiome data exploration as well as help uncover potential disease subgroups along with their potential associated biomarkers.”
Additionally, the method can be used to treat other diseases including diabetes, Chron’s disease, and ulcerative colitis which are believed to be related to the gut microbiome.
Hopefully, explainable AI will unveil additional secrets about the gut microbiome in the near future.
Source:
Journal reference:
Rynazal, R., et al. (2023). Leveraging explainable AI for gut microbiome-based colorectal cancer classification. Genome Biology. doi.org/10.1186/s13059-023-02858-4