Reviewed by Danielle Ellis, B.Sc.Jul 24 2023
Sensing and communicating with the extracellular environment are both aspects of transmembrane signaling. An important mechanism known as autoproteolysis, which is essential to many cellular processes, is a component of transmembrane signaling.
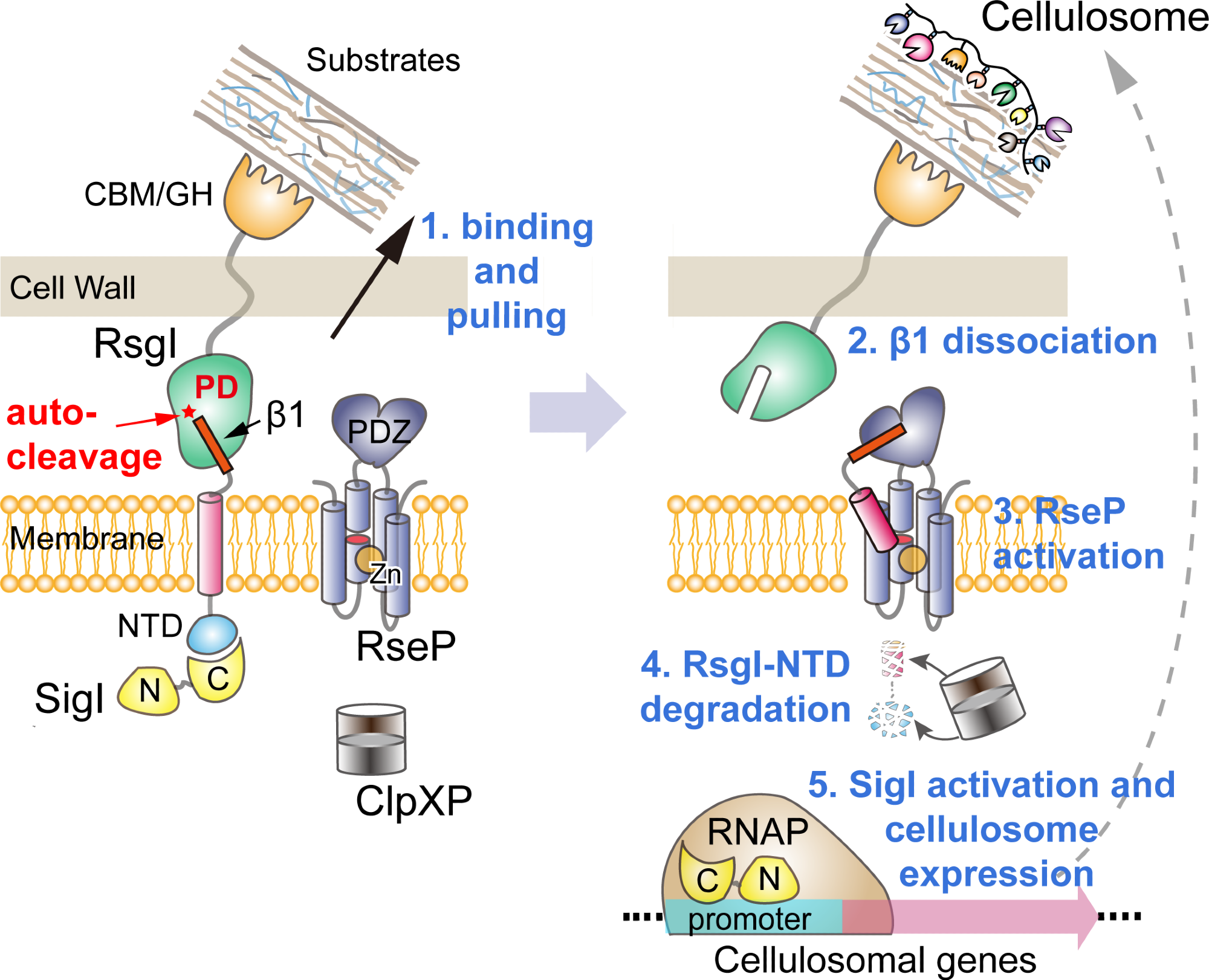 Proposed model of transmembrane signal transduction of RsgI in C. thermocellum. Image Credit: CHEN Chao and FEGN Yingang
Proposed model of transmembrane signal transduction of RsgI in C. thermocellum. Image Credit: CHEN Chao and FEGN Yingang
In this procedure, a peptide, which is a chain of amino acids, cleaves the chain at a certain point. Although a distinct, functioning autoproteolytic process for transmembrane signal transduction has not yet been observed in bacteria, this is typical of eukaryotes.
Researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) have now found evidence of this signaling process in the bacterial species Clostridium thermocellum (C. thermocellum), which was previously believed to be almost entirely unique to eukaryotes.
Researchers discovered an autoproteolytic effect in the bacterium C. thermocellum that is crucial for triggering later transmembrane signaling, comparable to the well-known autoproteolysis process seen in eukaryotes.
Image Credit: Kateryna Kon/Shutterstock.com
On July 7th, 2023, the study was published in Science Advances.
Not only is this a novel observation, but the presence of a conserved site for the automatic cleavage of the amino acid chain suggests a more unique and complex world of bacterial signaling than many would've thought possible.”
Dr Chao Chen, Study First Author and Associate Professor, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
Prokaryotes seldom report autoproteolysis for transmembrane signaling, and it is typically reserved for protease maturation. It appears to be a crucial component of signal transmission in the case of C. thermocellum, a bacterium that feeds on tough plant material (lignocellulolytic).
Asparagine-proline, a conserved amino acid sequence, is used in the process, which takes place between the “in-between” regions of the inner cytoplasmic membrane and the bacterial outer membrane known as the periplasm.
The anti- σfactor “RsgI” a protein that senses and transmits signals to cells, contains this conserved sequence. It also works to inhibit the actions of the σ-factor “SigI” which initiates the transcription of RNA from a DNA template and is responsible for transcribing specific genes.
Lignocellulolytic bacteria, such as the one used in this study, C. thermocellum, have extracellular enzyme complexes called “cellulosomes,” which are the targets regulated by numerous pairs of RsgI/SigI factors. The cellulosome functions to break down the hard plant materials the bacteria consume, such as cellulose, hemicellulose, and pectin.
As a unique class of σ/anti-σ factors, the functional mechanism of SigI/RsgI has still not been fully elucidated.”
Yingang Feng, Study Corresponding Author and Professor, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
Given the unique autoproteolytic mechanism observed in C. thermocellum, it is reasonable to assume that more research needs to be done to understand how this autoproteolysis functions in lignocellulolytic bacteria, particularly when it comes to the cellulosome and the associated regulatory actions carried out by anti-σ/σ factors, such as the binding of RNA polymerase to the promoter region to start transcription.
Source:
Journal reference:
Chen, C., et al. (2023). Essential autoproteolysis of bacterial anti-σ factor RsgI for transmembrane signal transduction. Science Advances. doi.org/10.1126/sciadv.adg4846