An international team of researchers has released the initial all-encompassing comparative analysis of the pan-genome of lactic acid bacteria (LAB), a group of microorganisms crucial to both natural ecosystems and the food industry.
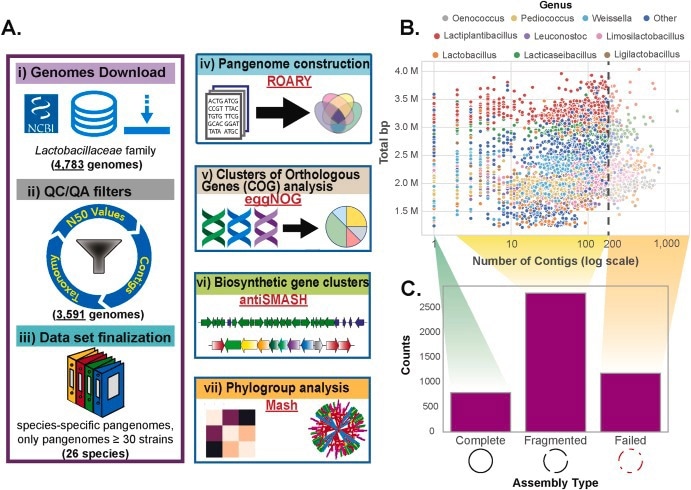
Image Credit: The Novo Nordisk Foundation Center for Biosustainability.
The investigation was conducted by scientists hailing from the Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain) and the University of California, San Diego.
Mapping the Genetic Landscape of Lactic Acid Bacteria
This groundbreaking study signifies a pivotal advancement in comprehending the genetic potential of 26 lactic acid bacteria (LAB) species. It is the first-ever comprehensive analysis of its kind encompassing the entire LAB family.
By examining more than 2400 publicly accessible genomes known for their high quality, the researchers effectively mapped the functional genetic capabilities, metabolic pathways, and biosynthetic gene clusters of individual strains spanning 26 species within the Lactobacillaceae family.
The researchers devised an innovative, all-encompassing workflow for large-scale pangenome analysis, which incorporated crucial stages like data curation, taxonomic categorization, phylogroup identification, pangenome reconstruction, functional annotations, and genome mining.
This advanced computational approach led to a deeper comprehension of the functional distinctions existing among various LAB species, surpassing previous endeavors that primarily concentrated on individual species or metagenome investigations.
In mapping the genetic landscape of LAB, we have cracked open the door to a treasure trove of novel possibilities represented by phylogenetic groups and individual strains for future biotechnological applications.”
Omkar Satyavan Mohite, Study Lead Author and Postdoc, Novo Nordisk Foundation Center for Biosustainability
Can Drive Advancement in the Food Industry and Healthcare
The results of this pioneering research extend beyond the realm of microbial data science, as they have practical applications in the real world.
We've laid the groundwork for everything from species identification and functional studies to phylogenetic research and biotechnological innovations. This work can drive advancements in various sectors, including the food industry, healthcare, and environmental sciences.”
Akanksha Rajput, Study Lead Author and Postdoctoral Researcher, University of California, San Diego
This research holds particular relevance for professionals and researchers in academic, industrial, medical, and environmental fields, all of whom can employ these data-driven insights to prioritize strains with specific genetic backgrounds.
The researchers anticipate that future studies will leverage pan-genomic characteristics as a fundamental tool to delve deeper into bacterial evolution, metabolic pathways, and sequence-level variations. The integrated pangenome analysis workflow developed here could serve as a critical platform for further explorations into other microbial groups with industrial significance.
Source:
Journal reference:
Rajput, A., et al. (2023). Pangenome analysis reveals the genetic basis for taxonomic classification of the Lactobacillaceae family. Food Microbiology. doi.org/10.1016/j.fm.2023.104334.