Reviewed by Danielle Ellis, B.Sc.Nov 22 2023
A recent examination has elucidated the molecular intricacies implicated in the sensory, trapping, and consumption processes of the carnivorous fungus Arthrobotrys oligospora when preying on worms.
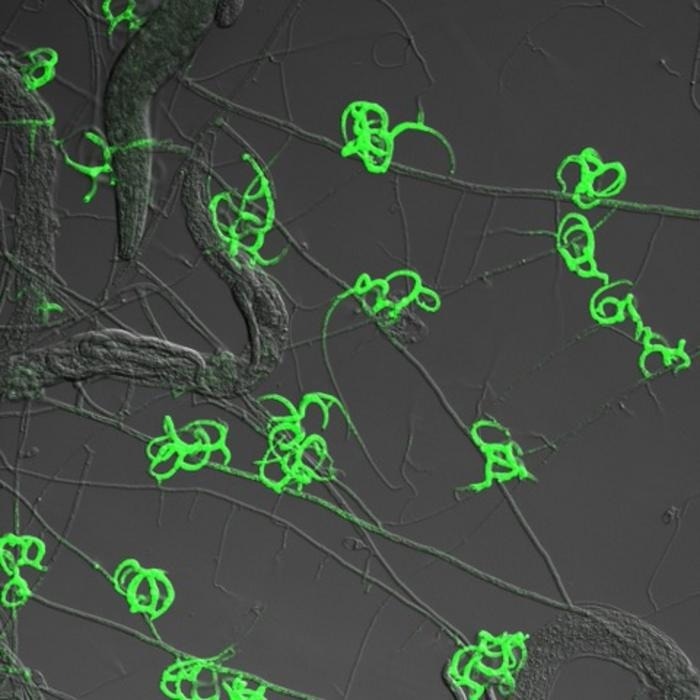 Glowing traps. Image Credit: Hung-Che Lin.
Glowing traps. Image Credit: Hung-Che Lin.
Presented by Hung-Che Lin and colleagues from Academia Sinica in Taipei, Taiwan, the revelations are featured in the open-access journal PLOS Biology on November 21st, 2023.
Arthrobotrys oligospora typically derives nourishment from decomposing organic matter; however, scarcity of nutrients and the proximity of worms can induce the fungus to construct traps for capturing and consuming them.
A. oligospora is among numerous fungi species capable of trapping and devouring minute animals. Previous studies have shed light on aspects of the biological dynamics in this predator-prey relationship, such as certain genes implicated in trap formation by A. oligospora. However, the finer molecular intricacies of the process have largely remained enigmatic.
To enhance comprehension, Lin and colleagues conducted a series of laboratory experiments exploring the genes and processes active during various stages of A. oligospora predation on the nematode worm species Caenorhabditis elegans.
Much of this analysis relied on RNAseq, a technique offering insights into the activity levels of different A. oligospora genes over time. The research unveiled several biological processes appearing pivotal in A. oligospora predation.
Upon initially detecting a worm, the findings propose an increase in DNA replication and the synthesis of ribosomes, cellular structures responsible for protein assembly.
Subsequently, the activity of numerous genes encoding proteins supporting trap formation and function, such as secreted worm-adhesive proteins and a newly identified protein family termed “trap enriched proteins:” (TEP), is heightened.
Finally, as A. oligospora extends filamentous hyphae into the worm for digestion, there is an upsurge in the activity of genes encoding various enzymes, particularly metalloproteases within the protease group. Since proteases break down other proteins, these results suggest that A. oligospora utilizes proteases to aid in the digestion of worms.
These observations could serve as a fundamental groundwork for future investigations into the molecular mechanisms underpinning A. oligospora predation and other interactions within the fungal predator-prey paradigm.
The authors add, “Our comprehensive transcriptomics and functional analyses highlight the role of increased DNA replication, translation, and secretion in trap development and efficacy.”
“Furthermore, a gene family that is largely expanded in the genomes of nematode-trapping fungi were found to be enriched in traps and critical for trap adhesion to nematodes. These results furthered our understanding of the key processes required for fungal carnivory,” concluded the authors.
Source:
Journal reference:
Lin, H-C., et al. (2023) Key processes required for the different stages of fungal carnivory by a nematode-trapping fungus. PLOS Biology. doi.org/10.1371/journal.pbio.3002400.