Reviewed by Danielle Ellis, B.Sc.Jan 3 2024
Researchers at Scripps Research created a novel, high-resolution method for identifying putative treatment targets on proteins in living cells.
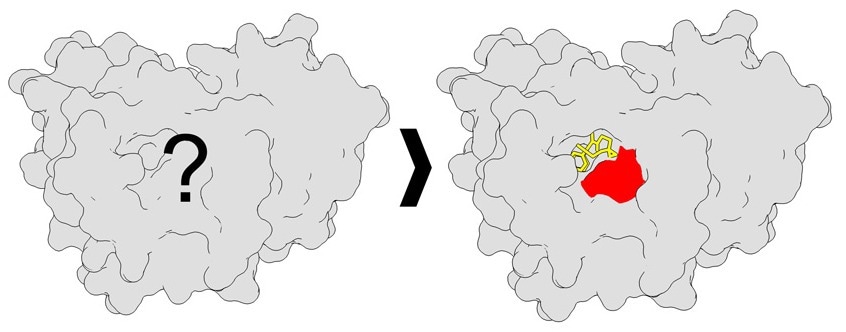 A protein with unknown binding sites (left), versus a protein showing high-resolution binding site mapping (right). Image Credit: The Scripps Research Institute.
A protein with unknown binding sites (left), versus a protein showing high-resolution binding site mapping (right). Image Credit: The Scripps Research Institute.
The discoveries may result in more specialized treatments for almost all diseases affecting humans.
Finding novel approaches to target proteins implicated in human diseases is a top concern for numerous researchers worldwide. Finding a way to change these proteins' functions, though, can be challenging, particularly in living cells. Now, researchers at Scripps Research have created a novel technique to study how proteins interact with small molecules that resemble drugs in human cells, providing important insights into possible therapeutic targets.
Image Credit: Billion Photos/Shutterstock.com
The strategy, which was published on January 2nd, 2024 in Nature Chemical Biology, combines analytical methods with chemistry to identify the precise locations where proteins and small molecules bind. In the end, this approach might result in the creation of medicines that are more efficient and precisely targeted.
Our new technology could be used to find new druggable sites on proteins for any human disease, from cancer to Alzheimer’s disease. We’re unrestricted in how this could be used. Our work has the potential to usher in a whole new way of drug discovery."
Christopher Parker PhD, Senior Author and Associate Professor, Department of Chemistry, The Scripps Research Institute
In order to create efficient treatments for a variety of human illnesses, the Parker lab seeks to understand how proteins function in each type of human cell. In this study, Parker and colleagues developed a novel technique for analyzing how proteins interact with small molecules in living cells, building on early work in the lab of Benjamin Cravatt, PhD, a Professor at Scripps Research.
To gain a deeper understanding of how these proteins interact with small molecules at a much higher resolution than before, the team devised an analytical method. The team accomplished this by using chemical probes known as photo affinity probes, which are molecules that, when exposed to light, become active and enable the probes to bind and capture proteins.
The Parker team determined the locations on proteins where small molecules could attach and bind by collecting data from the interactions of proteins with photo affinity probes. In essence, the team discovered more than a thousand novel locks - binding sites on the proteins - and corresponding keys, or small molecules, the great majority of which were previously unreported new locations for small-molecule binding. The team also discovered novel binding site characteristics, like altered shapes.
Identifying these specific binding sites will help scientists design new molecules that fit these pockets even better, potentially leading to more effective therapeutics."
Jacob M. Wozniak, Study Co-First Author and Former Postdoctoral Fellow, The Scripps Research Institute
The other co-first author of the paper was Weichao Li, PhD, a research associate also in the Parker lab.
The authors then modeled how specific molecules might bind to these proteins using the abundance of data in this study and working with co-author Stefano Forli, PhD, Associate Professor in the Department of Integrative Structural and Computational Biology. With the help of this information library, medicines with more focused interactions with proteins could be created.
Our new process reveals additional opportunities for therapeutic intervention and discovery in human cells. Next, we plan to use this technology to target proteins relevant for autoimmune diseases and cancer."
Christopher Parker PhD, Study Senior Author and Associate Professor, Department of Chemistry, The Scripps Research Institute
Source:
Journal reference:
Wozniak, M, J., et al. (2023). Enhanced mapping of small-molecule binding sites in cells. Nature Chemical Biology. doi/s41589-023-01514-z