Researchers from the Universities of Birmingham and Nottingham have uncovered how naturally occurring antimicrobial predatory bacteria, known as Bdellovibrio bacterivorous, produce fiber-like proteins on their surface to entice prey in a study published on January 4th, 2024, in Nature Microbiology.
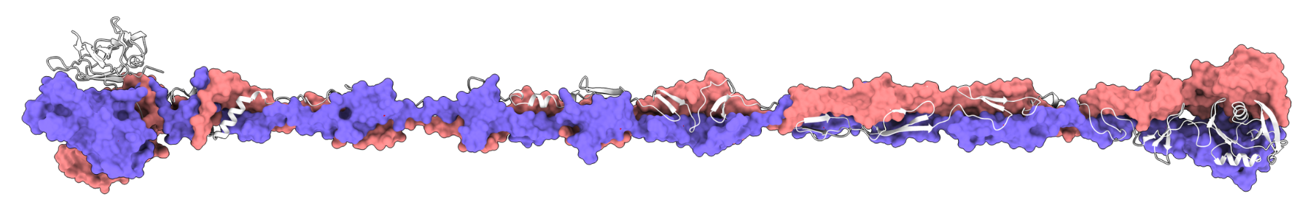 Bd2740 is a MAT fiber within the Bdellovibrio bacteriovorus genome. The bulb on the right side of the fiber is the part that attaches to Bdellvibrio's prey. Image Credit: University of Birmingham
Bd2740 is a MAT fiber within the Bdellovibrio bacteriovorus genome. The bulb on the right side of the fiber is the part that attaches to Bdellvibrio's prey. Image Credit: University of Birmingham
Thanks to this discovery, scientists may be able to employ these predators to target and eliminate harmful bacteria that harm the environment, healthcare, and food spoiling. The Wellcome Trust Investigator in Science Award (209437/Z/17/Z) provided funding for the study.
Since the 1960s, Bdellovibrio bacterivorous has been known to hunt and kill other bacteria by entering the target cells and eating them from the inside before later bursting out. The question that had stumped scientists was, ‘How do these cells make a firm attachment when we know how varied their bacterial targets are?’”
Andrew Lovering, Professor, Structural Biology, University of Birmingham
For nearly 15 years, Professor Lovering and Professor Liz Sockett of the University of Nottingham School of Life Sciences have worked together in this area. The discovery that the Bdellovibrio predators lay down a strong vesicle-a "pinched-off" portion of the predator cell envelope—when they invade their prey—was made by Ph.D. Student Asmaa Al-Bayati and Undergraduate Student Sam Greenwood in the Sockett lab.
The vesicle creates a kind of airlock or keyhole allowing Bdellovibrio entry into the prey cell. We were then able to isolate this vesicle from the dead prey, which is a first in this field. The vesicle was analyzed to reveal the tools used during the preceding event of predator/prey contact. We thought of it as a bit like a locksmith leaving the pick, or key, as evidence in the keyhole.”
Liz Sockett, Professor, School of Life Sciences, University of Nottingham
Liz Sockett explained, “By looking at the vesicle contents, we discovered that because Bdellovibrio doesn’t know which bacteria it will meet, it deploys a range of similar prey recognition molecules on its surface, creating lots of different ‘keys’ to ‘unlock’ lots of different types of prey.”
After that, the scientists examined each molecule separately and found that they formed lengthy fibers that were around 10 times longer than typical globular proteins. This enables them to function remotely and "feel" for potential prey in the surrounding area.
The labs counted 21 distinct fibers in total. Dr. Simon Caulton, Dr. Carey Lambert, and Dr. Jess Tyson conducted research on how they functioned at the molecular and cellular levels. Paul Radford and Rob Till used fiber gene engineering to support them.
Next, the group tried to connect a specific fiber to a specific prey-surface chemical. Determining which fiber corresponds to which prey may facilitate an engineering strategy in which customized predators target various bacterial species.
Professor Lovering continued, “Because the predator strain, we were looking at comes from the soil, it has a wide killing range, making this identification of these fiber and prey pairs very difficult. However, on the fifth attempt to find the partners, we discovered a chemical signature outside the prey bacteria that was a tight fit to the fiber tip. This is the first time a feature of Bdellovibrio has been matched to prey selection.”
Researchers studying this area will now be able to question which fiber set the various predators they research employ and perhaps even link these findings to particular prey. Gaining more knowledge about these predatory bacteria may make it possible to use them as antibiotics or to eradicate hazardous or food-degrading bacteria from the environment.
Professor Lovering concluded, “We know that these bacteria can be helpful, and by fully understanding how they operate and find their prey, it opens up a world of new discoveries and possibilities.”
Source:
Journal reference:
Caulton, S.G., et al., (2024). Bdellovibrio bacteriovorus uses chimeric fibre proteins to recognize and invade a broad range of bacterial hosts. Nature Microbiology. doi.org/10.1038/s41564-023-01552-2.